SIGLEC10+ macrophages drive gastric cancer progression by suppressing CD8+ T cell function
- PMID: 37432407
- PMCID: PMC10992087
- DOI: 10.1007/s00262-023-03488-2
SIGLEC10+ macrophages drive gastric cancer progression by suppressing CD8+ T cell function
Abstract
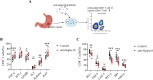
Existing immune checkpoint inhibitors focus on activating T cells and show limited effectiveness in gastric cancer (GC). SIGLEC10 is identified as a novel tumor-associated macrophage-related immune checkpoint in other cancer types. However, its immunosuppressive role and clinical significance in GC remain unclear. In this study, we find a dominant expression of SIGLEC10 on CD68+ macrophages in GC. SIGLEC10 can suppress the proliferation and function of tumor-infiltrating CD8+ T cells in vitro via the Akt/P38/Erk signaling pathway. Furthermore, in ex vivo and in vivo models, SIGLEC10 blockade promotes CD8+ T cell effector function. Finally, SIGLEC10+ macrophages are positively correlated with the adverse prognosis of GC. Our study highlights that SIGLEC10 directly suppresses T cell function and serves as a promising target for immunotherapy and suggests SIGLEC10+ macrophages as a novel potential predictor of the clinical prognosis of GC.
Keywords: CD68; CD8+ T cell; Gastric cancer; Macrophage; SIGLEC10.
© 2023. The Author(s), under exclusive licence to Springer-Verlag GmbH Germany, part of Springer Nature.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

