Stacking Interactions and Flexibility of Human Telomeric Multimers
- PMID: 37432645
- PMCID: PMC10375521
- DOI: 10.1021/jacs.3c04810
Stacking Interactions and Flexibility of Human Telomeric Multimers
Abstract
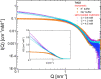
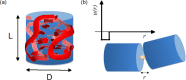
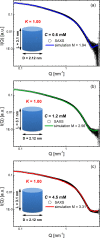
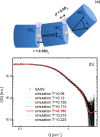
G-quadruplexes (G4s) are helical four-stranded structures forming from guanine-rich nucleic acid sequences, which are thought to play a role in cancer development and malignant transformation. Most current studies focus on G4 monomers, yet under suitable and biologically relevant conditions, G4s undergo multimerization. Here, we investigate the stacking interactions and structural features of telomeric G4 multimers by means of a novel low-resolution structural approach that combines small-angle X-ray scattering (SAXS) with extremely coarse-grained (ECG) simulations. The degree of multimerization and the strength of the stacking interaction are quantitatively determined in G4 self-assembled multimers. We show that self-assembly induces a significant polydispersity of the G4 multimers with an exponential distribution of contour lengths, consistent with a step-growth polymerization. On increasing DNA concentration, the strength of the stacking interaction between G4 monomers increases, as well as the average number of units in the aggregates. We utilized the same approach to explore the conformational flexibility of a model single-stranded long telomeric sequence. Our findings indicate that its G4 units frequently adopt a beads-on-a-string configuration. We also observe that the interaction between G4 units can be significantly affected by complexation with benchmark ligands. The proposed methodology, which identifies the determinants that govern the formation and structural flexibility of G4 multimers, may be an affordable tool aiding in the selection and design of drugs that target G4s under physiological conditions.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Neidle S. Quadruplex nucleic acids as targets for anticancer therapeutics. Nat. Rev. Chem. 2017, 1, 0041.10.1038/s41570-017-0041. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

