Dermokine mutations contribute to epithelial-mesenchymal transition and advanced melanoma through ERK/MAPK pathways
- PMID: 37432950
- PMCID: PMC10335698
- DOI: 10.1371/journal.pone.0285806
Dermokine mutations contribute to epithelial-mesenchymal transition and advanced melanoma through ERK/MAPK pathways
Retraction in
-
Retraction: Dermokine mutations contribute to epithelial-mesenchymal transition and advanced melanoma through ERK/MAPK pathways.PLoS One. 2024 Mar 26;19(3):e0300807. doi: 10.1371/journal.pone.0300807. eCollection 2024. PLoS One. 2024. PMID: 38530787 Free PMC article. No abstract available.
Abstract
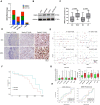
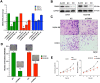
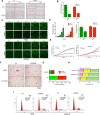
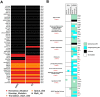
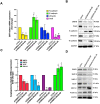
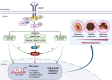
To discover vulnerabilities associated with dermokine (DMKN) as a new trigger of the epithelial-mesenchymal transition (EMT) -driven melanoma, we undertook a genome-wide genetic screening using transgenic. Here, we showed that DMKN expression could be constitutively increased in human malignant melanoma (MM) and that this correlates with poor overall survival in melanoma patients, especially in BRAF-mutated MM samples. Furthermore, in vitro, knockdown of DMKN inhibited the cell proliferation, migration, invasion, and apoptosis of MM cancer cells by the activation of ERK/MAPK signaling pathways and regulator of STAT3 in downstream molecular. By interrogating the in vitro melanoma dataset and characterization of advanced melanoma samples, we found that DMKN downregulated the EMT-like transcriptional program by disrupting EMT cortical actin, increasing the expression of epithelial markers, and decreasing the expression of mesenchymal markers. In addition, whole exome sequencing was presented with p.E69D and p.V91A DMKN mutations as a novel somatic loss of function mutations in those patients. Moreover, our purposeful proof-of-principle modeled the interaction of ERK with p.E69D and p.V91A DMKN mutations in the ERK-MAPK kinas signaling that may be naturally associated with triggering the EMT during melanomagenesis. Altogether, these findings provide preclinical evidence for the role of DMKN in shaping the EMT-like melanoma phenotype and introduced DMKN as a new exceptional responder for personalized MM therapy.
Copyright: © 2023 Ma et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Overexpression of Dermokine-α enhances the proliferation and epithelial-mesenchymal transition of pancreatic tumor cells.Cell Signal. 2022 Nov;99:110439. doi: 10.1016/j.cellsig.2022.110439. Epub 2022 Aug 15. Cell Signal. 2022. PMID: 35981655
-
Comparative mRNA/micro-RNA co-expression network drives melanomagenesis by promoting epithelial-mesenchymal transition and vasculogenic mimicry signaling.Transl Oncol. 2021 Dec;14(12):101237. doi: 10.1016/j.tranon.2021.101237. Epub 2021 Oct 6. Transl Oncol. 2021. PMID: 34626953 Free PMC article.
-
Dermokine contributes to epithelial-mesenchymal transition through increased activation of signal transducer and activator of transcription 3 in pancreatic cancer.Cancer Sci. 2017 Nov;108(11):2130-2141. doi: 10.1111/cas.13347. Epub 2017 Sep 15. Cancer Sci. 2017. PMID: 28795470 Free PMC article.
-
Potential therapeutic targets of epithelial-mesenchymal transition in melanoma.Cancer Lett. 2017 Apr 10;391:125-140. doi: 10.1016/j.canlet.2017.01.029. Epub 2017 Jan 25. Cancer Lett. 2017. PMID: 28131904 Free PMC article. Review.
-
Epithelial-to-mesenchymal-like transition events in melanoma.FEBS J. 2022 Mar;289(5):1352-1368. doi: 10.1111/febs.16021. Epub 2021 May 30. FEBS J. 2022. PMID: 33999497 Review.
Cited by
-
Characterization of two melanoma cell lines resistant to BRAF/MEK inhibitors (vemurafenib and cobimetinib).Cell Commun Signal. 2024 Aug 23;22(1):410. doi: 10.1186/s12964-024-01788-3. Cell Commun Signal. 2024. PMID: 39175042 Free PMC article.
-
Retraction: Dermokine mutations contribute to epithelial-mesenchymal transition and advanced melanoma through ERK/MAPK pathways.PLoS One. 2024 Mar 26;19(3):e0300807. doi: 10.1371/journal.pone.0300807. eCollection 2024. PLoS One. 2024. PMID: 38530787 Free PMC article. No abstract available.
-
Skin biophysical parameters and serum dermokine levels in airline pilots: a comparative study with office workers.Postepy Dermatol Alergol. 2023 Dec;40(6):757-761. doi: 10.5114/ada.2023.132262. Epub 2024 Jan 8. Postepy Dermatol Alergol. 2023. PMID: 38282882 Free PMC article.
-
How Protein Depletion Balances Thrombosis and Bleeding Risk in the Context of Platelet's Activatory and Negative Signaling.Int J Mol Sci. 2024 Sep 17;25(18):10000. doi: 10.3390/ijms251810000. Int J Mol Sci. 2024. PMID: 39337488 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

