Tumor monocyte content predicts immunochemotherapy outcomes in esophageal adenocarcinoma
- PMID: 37433281
- PMCID: PMC11913779
- DOI: 10.1016/j.ccell.2023.06.006
Tumor monocyte content predicts immunochemotherapy outcomes in esophageal adenocarcinoma
Abstract
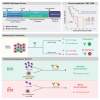
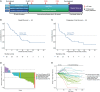
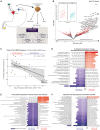
For inoperable esophageal adenocarcinoma (EAC), identifying patients likely to benefit from recently approved immunochemotherapy (ICI+CTX) treatments remains a key challenge. We address this using a uniquely designed window-of-opportunity trial (LUD2015-005), in which 35 inoperable EAC patients received first-line immune checkpoint inhibitors for four weeks (ICI-4W), followed by ICI+CTX. Comprehensive biomarker profiling, including generation of a 65,000-cell single-cell RNA-sequencing atlas of esophageal cancer, as well as multi-timepoint transcriptomic profiling of EAC during ICI-4W, reveals a novel T cell inflammation signature (INCITE) whose upregulation correlates with ICI-induced tumor shrinkage. Deconvolution of pre-treatment gastro-esophageal cancer transcriptomes using our single-cell atlas identifies high tumor monocyte content (TMC) as an unexpected ICI+CTX-specific predictor of greater overall survival (OS) in LUD2015-005 patients and of ICI response in prevalent gastric cancer subtypes from independent cohorts. Tumor mutational burden is an additional independent and additive predictor of LUD2015-005 OS. TMC can improve patient selection for emerging ICI+CTX therapies in gastro-esophageal cancer.
Keywords: cell type deconvolution; esophageal cancer; immune checkpoint inhibitors; immune priming; immunochemotherapy; molecular profiling; predictive biomarkers; single-cell RNA-sequencing atlas; tumor associated monocytes; tumor mutational burden.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests S.L.: consulting fees, honoraria, travel/accommodation or research funding (Sanofi, GLG Consulting, Rejuversen, Eisai, Prosigna, Roche, Pfizer, Novartis, Shionogi, Synthon, CRUK, Boehringer Ingelheim, Piqur Therapeutics, AstraZeneca, Carrick Therapeutics, Merck KGaA) and previous employment by Pfizer. A.R.: stock ownership (Amgen, Immunogen). I.K: honoraria, travel/accommodation (BMS , Delcath Inc, Immunocore, Pierre Fabre, Genentech, Merck Serono, Takeda Pharmaceuticals Int.). B.J.V.D.E.: consulting and ownership interests (iTeos Therapeutics, Oncorus, Amgen, Vaccitech). M.R.M.: grants or personal fees (AstraZeneca, Roche, G.S.K., Novartis, Immunocore, BMS, Pfizer, Merck/MSD, Regeneron, BiolineRx, Replimune, Kineta, Silicon Therapeutics and GRAIL). T.M.C.: founder, employee, and shareholder (Cleancard). X.L.: consulting (SimCell). A provisional patent related to applications of the INCITE signature has been filed. No other authors declare competing interests.
Figures





Comment in
-
Precise patient stratification in esophageal cancer: Biomarkers for immunochemotherapy.Cancer Cell. 2023 Jul 10;41(7):1199-1201. doi: 10.1016/j.ccell.2023.06.004. Cancer Cell. 2023. PMID: 37433280
References
-
- Xin Yu J., Hubbard-Lucey V.M., Tang J. Immuno-oncology drug development goes global. Nat. Rev. Drug Discov. 2019;18:899–900. - PubMed
-
- Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 2018;68:394–424. - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

