Genome analysis of the novel putative rotavirus species K
- PMID: 37433351
- PMCID: PMC10410577
- DOI: 10.1016/j.virusres.2023.199171
Genome analysis of the novel putative rotavirus species K
Abstract
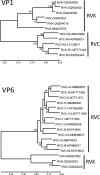
Rotaviruses are causative agents of diarrhea in humans and animals. Currently, the species rotavirus A-J (RVA-RVJ) and the putative species RVK and RVL are defined, mainly based on their genome sequence identities. RVK strains were first identified in 2019 in common shrews (Sorex aranaeus) in Germany; however, only short sequence fragments were available so far. Here, we analyzed the complete coding regions of strain RVK/shrew-wt/GER/KS14-0241/2013, which showed highest sequence identities with RVC. The amino acid sequence identity of VP6, which is used for rotavirus species definition, reached only 51% with other rotavirus reference strains thus confirming classification of RVK as a separate species. Phylogenetic analyses for the deduced amino acid sequences of all 11 virus proteins showed, that for most of them RVK and RVC formed a common branch within the RVA-like phylogenetic clade. Only the tree for the highly variable NSP4 showed a different branching; however, with very low bootstrap support. Comparison of partial nucleotide sequences of other RVK strains from common shrews of different regions in Germany indicated a high degree of sequence variability (61-97% identity) within the putative species. These RVK strains clustered separately from RVC genotype reference strains in phylogenetic trees indicating diversification of RVK independent from RVC. The results indicate that RVK represents a novel rotavirus species, which is most closely related to RVC.
Keywords: Genome sequence; Phylogeny; Rotavirus; Shrew; Taxonomy.
Copyright © 2023. Published by Elsevier B.V.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

