Translational regulation by uORFs and start codon selection stringency
- PMID: 37433636
- PMCID: PMC10393191
- DOI: 10.1101/gad.350752.123
Translational regulation by uORFs and start codon selection stringency
Abstract
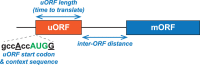
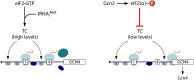
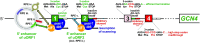
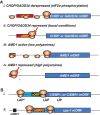
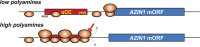
In addition to the main, protein-coding, open reading frame (mORF), many eukaryotic mRNAs contain upstream ORFs (uORFs) initiated at AUG or near-cognate codons residing 5' of the mORF start site. Whereas translation of uORFs generally represses translation of the mORFs, a subset of uORFs serves as a nexus for regulating translation of the mORF. In this review, we summarize the mechanisms by which uORFs can repress or stimulate mRNA translation, highlight uORF-mediated translational repression involving ribosome queuing, and critically evaluate recently described alternatives to the delayed reinitiation model for uORF-mediated regulation of the GCN4/ATF4 mRNAs.
Keywords: ATF4; GCN4; eIF2 phosphorylation; ribosome queuing; stringency; uORF.
Published by Cold Spring Harbor Laboratory Press.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
