Expression and possible functions of a horizontally transferred glycosyl hydrolase gene, GH6-1, in Ciona embryogenesis
- PMID: 37434168
- PMCID: PMC10334666
- DOI: 10.1186/s13227-023-00215-x
Expression and possible functions of a horizontally transferred glycosyl hydrolase gene, GH6-1, in Ciona embryogenesis
Abstract
Background: The Tunicata or Urochordata is the only animal group with the ability to synthesize cellulose directly and cellulose is a component of the tunic that covers the entire tunicate body. The genome of Ciona intestinalis type A contains a cellulose synthase gene, CesA, that it acquired via an ancient, horizontal gene transfer. CesA is expressed in embryonic epidermal cells and functions in cellulose production. Ciona CesA is composed of both a glycosyltransferase domain, GT2, and a glycosyl hydrolase domain, GH6, which shows a mutation at a key position and seems functionless. Interestingly, the Ciona genome contains a glycosyl hydrolase gene, GH6-1, in which the GH6 domain seems intact. This suggests expression and possible functions of GH6-1 during Ciona embryogenesis. Is GH6-1 expressed during embryogenesis? If so, in what tissues is the gene expressed? Does GH6-1 serve a function? If so, what is it? Answers to these questions may advance our understanding of evolution of this unique animal group.
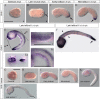
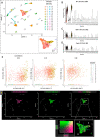
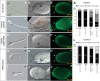
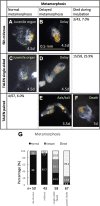
Results: Quantitative reverse transcription PCR and in situ hybridization revealed that GH6-1 is expressed in epidermis of tailbud embryos and in early swimming larvae, a pattern similar to that of CesA. Expression is downregulated at later stages and becomes undetectable in metamorphosed juveniles. The GH6-1 expression level is higher in the anterior-trunk region and caudal-tip regions of late embryos. Single-cell RNA sequencing analysis of the late tailbud stage showed that cells of three clusters with epidermal identity express GH6-1, and that some of them co-express CesA. TALEN-mediated genome editing was used to generate GH6-1 knockout Ciona larvae. Around half of TALEN-electroporated larvae showed abnormal development of adhesive papillae and altered distribution of surface cellulose. In addition, three-fourths of TALEN-electroporated animals failed to complete larval metamorphosis.
Conclusions: This study showed that tunicate GH6-1, a gene that originated by horizontal gene transfer of a prokaryote gene, is recruited into the ascidian genome, and that it is expressed and functions in epidermal cells of ascidian embryos. Although further research is required, this observation demonstrates that both CesA and GH6-1 are involved in tunicate cellulose metabolism, impacting tunicate morphology and ecology.
Keywords: Expression and function in Ciona embryogenesis; Glycosyl hydrolase family-6 GH6 gene; Horizontally transferred genes; Tunicates.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Phylogenetic Analyses of Glycosyl Hydrolase Family 6 Genes in Tunicates: Possible Horizontal Transfer.Genes (Basel). 2020 Aug 13;11(8):937. doi: 10.3390/genes11080937. Genes (Basel). 2020. PMID: 32823766 Free PMC article.
-
Transposon-mediated insertional mutagenesis revealed the functions of animal cellulose synthase in the ascidian Ciona intestinalis.Proc Natl Acad Sci U S A. 2005 Oct 18;102(42):15134-9. doi: 10.1073/pnas.0503640102. Epub 2005 Oct 7. Proc Natl Acad Sci U S A. 2005. PMID: 16214891 Free PMC article.
-
Functional specialization of cellulose synthase genes of prokaryotic origin in chordate larvaceans.Development. 2010 May;137(9):1483-92. doi: 10.1242/dev.044503. Epub 2010 Mar 24. Development. 2010. PMID: 20335363
-
[Ascidian embryos: from the birth of experimental embryology to the analysis of gene regulatory networks].Med Sci (Paris). 2008 Mar;24(3):263-9. doi: 10.1051/medsci/2008243263. Med Sci (Paris). 2008. PMID: 18334174 Review. French.
-
Gene-regulatory networks in the Ciona embryos.Brief Funct Genomic Proteomic. 2009 Jul;8(4):250-5. doi: 10.1093/bfgp/elp018. Epub 2009 Jun 17. Brief Funct Genomic Proteomic. 2009. PMID: 19535506 Review.
Cited by
-
Molecular control of cellulosic fin morphogenesis in ascidians.BMC Biol. 2024 Apr 2;22(1):74. doi: 10.1186/s12915-024-01872-7. BMC Biol. 2024. PMID: 38561802 Free PMC article.
-
CRISPR/Cas9 Protocols for Disrupting Gene Function in the Non-vertebrate Chordate Ciona.Integr Comp Biol. 2024 Nov 21;64(5):1182-1193. doi: 10.1093/icb/icae108. Integr Comp Biol. 2024. PMID: 38982335 Free PMC article. Review.
References
-
- Satoh, N. 2014. Developmental Genomics of Ascidians, Hoboken, New Jersey, Wiley-Blackwell, 9781118656181.
LinkOut - more resources
Full Text Sources
Research Materials

