The Ephrin tyrosine kinase a3 (EphA3) is a novel mediator of RAGE-prompted motility of breast cancer cells
- PMID: 37434266
- PMCID: PMC10337103
- DOI: 10.1186/s13046-023-02747-5
The Ephrin tyrosine kinase a3 (EphA3) is a novel mediator of RAGE-prompted motility of breast cancer cells
Abstract
Background: The receptor for advanced glycation-end products (RAGE) and its ligands have been implicated in obesity and associated inflammatory processes as well as in metabolic alterations like diabetes. In addition, RAGE-mediated signaling has been reported to contribute to the metastatic progression of breast cancer (BC), although mechanistic insights are still required. Here, we provide novel findings regarding the transcriptomic landscape and the molecular events through which RAGE may prompt aggressive features in estrogen receptor (ER)-positive BC.
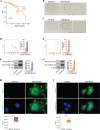
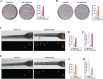
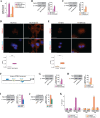
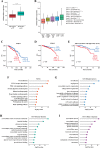
Methods: MCF7 and T47D BC cells stably overexpressing human RAGE were used as a model system to evaluate important changes like cell protrusions, migration, invasion and colony formation both in vitro through scanning electron microscopy, clonogenic, migration and invasion assays and in vivo through zebrafish xenografts experiments. The whole transcriptome of RAGE-overexpressing BC cells was screened by high-throughput RNA sequencing. Thereafter, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses allowed the prediction of potential functions of differentially expressed genes (DEGs). Flow cytometry, real time-PCR, chromatin immunoprecipitation, immunofluorescence and western blot assays were performed to investigate the molecular network involved in the regulation of a novel RAGE target gene namely EphA3. The clinical significance of EphA3 was explored in the TCGA cohort of patients through the survivALL package, whereas the pro-migratory role of EphA3 signaling was ascertained in both BC cells and cancer-associated fibroblasts (CAFs). Statistical analysis was performed by t-tests.
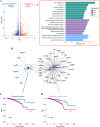
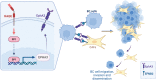
Results: RNA-seq findings and GSEA analysis revealed that RAGE overexpression leads to a motility-related gene signature in ER-positive BC cells. Accordingly, we found that RAGE-overexpressing BC cells exhibit long filopodia-like membrane protrusions as well as an enhanced dissemination potential, as determined by the diverse experimental assays. Mechanistically, we established for the first time that EphA3 signaling may act as a physical mediator of BC cells and CAFs motility through both homotypic and heterotypic interactions.
Conclusions: Our data demonstrate that RAGE up-regulation leads to migratory ability in ER-positive BC cells. Noteworthy, our findings suggest that EphA3 may be considered as a novel RAGE target gene facilitating BC invasion and scattering from the primary tumor mass. Overall, the current results may provide useful insights for more comprehensive therapeutic approaches in BC, particularly in obese and diabetic patients that are characterized by high RAGE levels.
Keywords: Breast cancer; Cancer-associated fibroblast (CAFs); EphA3; Receptor for advanced glycation end-products (RAGE).
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

