Blind fish have cells that see light
- PMID: 37434525
- PMCID: PMC10336380
- DOI: 10.1098/rspb.2023.0981
Blind fish have cells that see light
Abstract
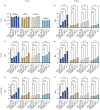
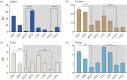
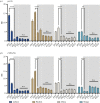
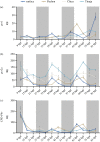
Most animals on earth have evolved under daily light-dark cycles and consequently possess a circadian clock which regulates much of their biology, from cellular processes to behaviour. There are however some animals that have invaded dark ecosystems and have adapted to an apparently arrhythmic environment. One such example is the Mexican blind cavefish Astyanax mexicanus, a species complex with over 30 different isolated cave types, including the founding surface river fish. These cavefish have evolved numerous fascinating adaptations to the dark, such as loss of eyes, reduced sleep phenotype and alterations in their clock and light biology. While cavefish are an excellent model for studying circadian adaptations to the dark, their rarity and long generational time makes many studies challenging. To overcome these limitations, we established embryonic cell cultures from cavefish strains and assessed their potential as tools for circadian and light experiments. Here, we show that despite originating from animals with no eyes, cavefish cells in culture are directly light responsive and show an endogenous circadian rhythm, albeit that light sensitivity is relatively reduced in cave strain cells. Expression patterns are similar to adult fish, making these cavefish cell lines a useful tool for further circadian and molecular studies.
Keywords: Astyanax mexicanus; biological clock; cavefish; cell culture; circadian; light biology.
Conflict of interest statement
We declare we have no competing interests.
Figures
References
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources

