Effect of temperature on betacyanins synthesis and the transcriptome of Suaeda salsa
- PMID: 37434603
- PMCID: PMC10330703
- DOI: 10.3389/fpls.2023.1203089
Effect of temperature on betacyanins synthesis and the transcriptome of Suaeda salsa
Abstract
Introduction: Suaeda salsa (Linn.) Pall. is an important tourist resource and ecological restoration species in coastal wetlands. Environmental factors such as low temperature, darkness, phytohormone, salt stress and seawater flflooding, and light can induce betalain synthesis in S. salsa, which plays an important role in plant adaptation to abiotic stress processes and in shaping the beautiful "red beach" landscape.
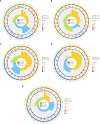
Methods: In this study, Illumina sequencing was used to profifile the transcriptome sequence (RNA-Seq) of S. salsa leaves at different temperatures (5° C, 10°C, 15°C, 20°C, 25°C, and 30°C) and to validate differentially expressed genes (DEGs) indicated by real-time PCR (RT-qPCR).
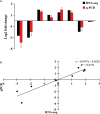
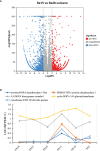
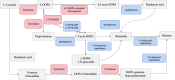
Results: The betacyanin content was highest in S. salsa leaves at 15°C. Transcription group data showed that compared to the control group (15°C), the "betacyanin biosynthesis pathway" was signifificantly enriched in the fifive different temperature groups. KEGG analysis showed that the DEGs were mainly involved in pathways of phenylpropanoid biosynthesis, carbon fifixation in photosynthetic organisms, flflavonoid biosynthesis, and betacyanin biosynthesis. Among the key enzymes involved in biosynthesis of betacyanin, genes for tyrosinase, CYP76AD1 and 4,5-DOPA dioxygenase were signifificantly upregulated and most abundantly expressed at 15°C. It is possible that the gene for betacyanin synthesis from S. salsa is primarily regulated by the MYB1R1 and MYB1 transcription factor. Four DEGs were randomly selected for quantitative PCR analysis, and DEG expression was generally consistent with the RNA-Seq data, verifying the validity of the transcriptome sequencing data.
Discussion: Relative to other temperatures, 15°C was optimum for S. salsa betacyanin synthesis, and this provides a theoretical reference for coastal wetland ecological remediation, reveals mechanisms of S. salsa discoloration, and further mines its potential application for landscape vegetation.
Keywords: Suaeda salsa; betacyanins; coastal wetlands; temperature; transcriptome.
Copyright © 2023 Li, He, Zhao, Liu, Liu, Ma, Shen and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Chen J., Xie F. F., Shah K., Chen C. B., Zeng J. M., Chen J. X., et al. . (2023). Identification of HubHLH family and key role of HubHLH159 in betalain biosynthesis by activating the transcription of HuADH1, HuCYP76AD1-1, and HuDODA1 in pitaya . Plant Sci. 328, 111595. doi: 10.1016/j.plantsci.2023.111595 - DOI - PubMed
-
- Cui M. Z. (2020). Biosynthesis of betacyanins in suaeda salsa under the combined stress of low nitrogen, low phosphorus and high salt and its effect on plant salt tolerance (Lanzhou (CN: Lan Zhou University; ).
LinkOut - more resources
Full Text Sources

