Screening of novel biomarkers for breast cancer based on WGCNA and multiple machine learning algorithms
- PMID: 37434679
- PMCID: PMC10331707
- DOI: 10.21037/tcr-23-3
Screening of novel biomarkers for breast cancer based on WGCNA and multiple machine learning algorithms
Abstract
Background: Breast cancer (BC) ranks first in incidence among women, with approximately 2 million new cases per year. Therefore, it is essential to investigate emerging targets for BC patients' diagnosis and prognosis.
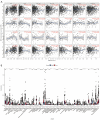
Methods: We analyzed gene expression data from 99 normal and 1,081 BC tissues in The Cancer Genome Atlas (TCGA) database. Differentially expressed genes (DEGs) were identified using "limma" R package, and relevant modules were chosen through Weighted Gene Coexpression Network Analysis (WGCNA). Intersection genes were obtained by matching DEGs to WGCNA module genes. Functional enrichment studies were performed on these genes using Gene Ontology (GO), Disease Ontology (DO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. Biomarkers were screened via Protein-Protein Interaction (PPI) networks and multiple machine-learning algorithms. The Gene Expression Profiling Interactive Analysis (GEPIA), The University of ALabama at Birmingham CANcer (UALCAN), and Human Protein Atlas (HPA) databases were employed to examine mRNA and protein expression of eight biomarkers. Kaplan-Meier mapper tool assessed their prognostic capabilities. Key biomarkers were analyzed via single-cell sequencing, and their relationship with immune infiltration was examined using Tumor Immune Estimation Resource (TIMER) database and "xCell" R package. Lastly, drug prediction was conducted based on the identified biomarkers.
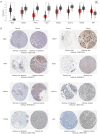
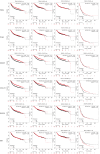
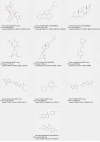
Results: We identified 1,673 DEGs and 542 important genes through differential analysis and WGCNA, respectively. Intersection analysis revealed 76 genes, which play significant roles in immune-related viral infection and IL-17 signaling pathways. DIX domain containing 1 (DIXDC1), Dual specificity phosphatase 6 (DUSP6), Pyruvate dehydrogenase kinase 4 (PDK4), C-X-C motif chemokine ligand 12 (CXCL12), Interferon regulatory factor 7 (IRF7), Integrin subunit alpha 7 (ITGA7), NIMA related kinase 2 (NEK2), and Nuclear receptor subfamily 3 group C member 1 (NR3C1) were selected as BC biomarkers using machine-learning algorithms. NEK2 was the most critical gene for diagnosis. Prospective drugs targeting NEK2 include etoposide and lukasunone.
Conclusions: Our study identified DIXDC1, DUSP6, PDK4, CXCL12, IRF7, ITGA7, NEK2, and NR3C1 as potential diagnostic biomarkers for BC, with NEK2 having the highest potential to aid in diagnosis and prognosis in clinical settings.
Keywords: Machine learning; NEK2; biomarkers; breast cancer (BC); weighted correlation network analysis.
2023 Translational Cancer Research. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-3/coif). The authors have no conflicts of interest to declare.
Figures




Similar articles
-
Elucidating the molecular and immune interplay between head and neck squamous cell carcinoma and diffuse large B-cell lymphoma through bioinformatics and machine learning.Transl Cancer Res. 2024 Nov 30;13(11):5725-5750. doi: 10.21037/tcr-24-1064. Epub 2024 Nov 21. Transl Cancer Res. 2024. PMID: 39697749 Free PMC article.
-
Coexpression Module Construction by Weighted Gene Coexpression Network Analysis and Identify Potential Prognostic Markers of Breast Cancer.Cancer Biother Radiopharm. 2022 Oct;37(8):612-623. doi: 10.1089/cbr.2020.3821. Epub 2020 Oct 14. Cancer Biother Radiopharm. 2022. PMID: 33052716
-
Screening of potential biomarkers in peripheral blood of patients with depression based on weighted gene co-expression network analysis and machine learning algorithms.Front Psychiatry. 2022 Oct 17;13:1009911. doi: 10.3389/fpsyt.2022.1009911. eCollection 2022. Front Psychiatry. 2022. PMID: 36325528 Free PMC article.
-
WGCNA combined with machine learning algorithms for analyzing key genes and immune cell infiltration in heart failure due to ischemic cardiomyopathy.Front Cardiovasc Med. 2023 Mar 17;10:1058834. doi: 10.3389/fcvm.2023.1058834. eCollection 2023. Front Cardiovasc Med. 2023. PMID: 37008314 Free PMC article.
-
Identification of biomarkers associated with pediatric asthma using machine learning algorithms: A review.Medicine (Baltimore). 2023 Nov 24;102(47):e36070. doi: 10.1097/MD.0000000000036070. Medicine (Baltimore). 2023. PMID: 38013370 Free PMC article. Review.
Cited by
-
Identification of biomarkers related to Escherichia coli infection for the diagnosis of gastrointestinal tumors applying machine learning methods.Heliyon. 2024 Nov 16;10(23):e40491. doi: 10.1016/j.heliyon.2024.e40491. eCollection 2024 Dec 15. Heliyon. 2024. PMID: 39654750 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous
