N-terminal lid swapping contributes to the substrate specificity and activity of thermophilic lipase TrLipE
- PMID: 37434709
- PMCID: PMC10332459
- DOI: 10.3389/fmicb.2023.1193955
N-terminal lid swapping contributes to the substrate specificity and activity of thermophilic lipase TrLipE
Abstract
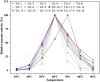
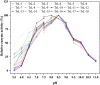
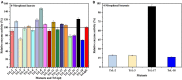
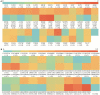
TrLipE is a thermophilic lipase that has potential commercial applications because of its catalytic ability under extreme conditions. Consistent with most lipases, the lid of TrLipE is located over the catalytic pocket, controls the substrate channel to the active center, and regulates the substrate specificity, activity, and stability of the enzyme through conformational changes. TrLipE from Thermomicrobium roseum has potential industrial applications, which is hindered by its weak enzymatic activity. Here, 18 chimeras (TrL1-TrL18) were reconstructed by N-terminal lid swapping between TrLipE and structurally similar enzymes. The results showed that the chimeras had a similar pH range and optimum pH as wild TrLipE but a narrower temperature range of 40-80°C, and TrL17 and the other chimeras showed lower optimum temperatures of 70°C and 60°C, respectively. In addition, the half-lives of the chimeras were lower than those of TrLipE under optimum temperature conditions. Molecular dynamics simulations indicated that chimeras had high RMSD, RMSF, and B-factor values. When p-nitrophenol esters with different chains were used as substrates, compared with TrLipE, most of the chimeras had a low Km and high kcat value. The chimeras TrL2, TrL3, TrL17, and TrL18 could specifically catalyze the substrate 4-nitrophenyl benzoate, with TrL17 showing the highest kcat/Km value of 363.88 ± 15.83 L⋅min-1⋅mmol-1. Mutants were then designed by investigating the binding free energies of TrL17 and 4-nitrophenyl benzoate. The results indicated that single, double, and triple substitution variants (M89W and I206N; E33W/I206M and M89W/I206M; and M89W/I206M/L21I and M89W/I206N/L21I, respectively) presented approximately 2- to 3-fold faster catalysis of 4-nitrophenyl benzoate than the wild TrL17. Our observations will facilitate the development of the properties and industrial applications of TrLipE.
Keywords: binding energy; lid swapping; molecular dynamics; substrate specificity; thermophilic lipase.
Copyright © 2023 Fang, Liu, Shi, Yang, Xin, Gu, Shi and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
N-terminal domain replacement changes an archaeal monoacylglycerol lipase into a triacylglycerol lipase.Biotechnol Biofuels. 2019 May 6;12:110. doi: 10.1186/s13068-019-1452-5. eCollection 2019. Biotechnol Biofuels. 2019. PMID: 31080517 Free PMC article.
-
Ancestral sequence reconstruction and spatial structure analysis guided alteration of longer-chain substrate catalysis for Thermomicrobium roseum lipase.Enzyme Microb Technol. 2022 May;156:109989. doi: 10.1016/j.enzmictec.2022.109989. Epub 2022 Jan 22. Enzyme Microb Technol. 2022. PMID: 35134708
-
Conversion of a Rhizopus chinensis lipase into an esterase by lid swapping.J Lipid Res. 2014 Jun;55(6):1044-51. doi: 10.1194/jlr.M043950. Epub 2014 Mar 26. J Lipid Res. 2014. PMID: 24670990 Free PMC article.
-
Cold active microbial lipases: some hot issues and recent developments.Biotechnol Adv. 2008 Sep-Oct;26(5):457-70. doi: 10.1016/j.biotechadv.2008.05.003. Epub 2008 May 18. Biotechnol Adv. 2008. PMID: 18571355 Review.
-
Protein engineering and applications of Candida rugosa lipase isoforms.Lipids. 2004 Jun;39(6):513-26. doi: 10.1007/s11745-004-1258-7. Lipids. 2004. PMID: 15554150 Review.
Cited by
-
Conserved specificity of extracellular wastewater peptidases revealed by multiplex substrate profiling by mass spectrometry.Environ Chem Lett. 2025;23(4):953-959. doi: 10.1007/s10311-025-01834-7. Epub 2025 Apr 12. Environ Chem Lett. 2025. PMID: 40453165 Free PMC article.
-
Enhancing the Hydrolytic Activity of a Lipase towards Larger Triglycerides through Lid Domain Engineering.Int J Mol Sci. 2023 Sep 6;24(18):13768. doi: 10.3390/ijms241813768. Int J Mol Sci. 2023. PMID: 37762071 Free PMC article.
References
-
- Almeida F., Travália B. M., Gonalves I. S., Forte M. B. S. (2021). Biodiesel production by lipase-catalyzed reactions: Bibliometric analysis and study of trends. Biofuel. Bioprod. Biorefin. 15 1141–1159.
-
- Anuar N. F. S. K., Wahab R. A., Huyop F., Amran S. I., Hamid A. A. A., Halim K. B. A., et al. (2021). Molecular docking and molecular dynamics simulations of a mutant Acinetobacter haemolyticus alkaline-stable lipase against tributyrin. J. Biomol. Struct. Dyn. 39 2079–2091. 10.1080/07391102.2020.1743364 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

