Diagnostic performance and clinical impact of blood metagenomic next-generation sequencing in ICU patients suspected monomicrobial and polymicrobial bloodstream infections
- PMID: 37434786
- PMCID: PMC10330723
- DOI: 10.3389/fcimb.2023.1192931
Diagnostic performance and clinical impact of blood metagenomic next-generation sequencing in ICU patients suspected monomicrobial and polymicrobial bloodstream infections
Abstract
Introduction: Early and effective application of antimicrobial medication has been evidenced to improve outcomes of patients with bloodstream infection (BSI). However, conventional microbiological tests (CMTs) have a number of limitations that hamper a rapid diagnosis.
Methods: We retrospectively collected 162 cases suspected BSI from intensive care unit with blood metagenomics next-generation sequencing (mNGS) results, to comparatively evaluate the diagnostic performance and the clinical impact on antibiotics usage of mNGS.
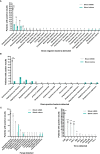
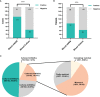
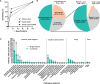
Results and discussion: Results showed that compared with blood culture, mNGS detected a greater number of pathogens, especially for Aspergillus spp, and yielded a significantly higher positive rate. With the final clinical diagnosis as the standard, the sensitivity of mNGS (excluding viruses) was 58.06%, significantly higher than that of blood culture (34.68%, P<0.001). Combing blood mNGS and culture results, the sensitivity improved to 72.58%. Forty-six patients had infected by mixed pathogens, among which Klebsiella pneumoniae and Acinetobacter baumannii contributed most. Compared to monomicrobial, cases with polymicrobial BSI exhibited dramatically higher level of SOFA, AST, hospitalized mortality and 90-day mortality (P<0.05). A total of 101 patients underwent antibiotics adjustment, among which 85 were adjusted according to microbiological results, including 45 cases based on the mNGS results (40 cases escalation and 5 cases de-escalation) and 32 cases on blood culture. Collectively, for patients suspected BSI in critical condition, mNGS results can provide valuable diagnostic information and contribute to the optimizing of antibiotic treatment. Combining conventional tests with mNGS may significantly improve the detection rate for pathogens and optimize antibiotic treatment in critically ill patients with BSI.
Keywords: blood culture; bloodstream infection; intensive care unit; metagenomics next generation sequencing; polymicrobial infection.
Copyright © 2023 Liu, Liu, Hu, Xu, Sun, Li, Zhang, Yang, Ma and Sun.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Performance of next-generation sequencing for diagnosis of blood infections by Klebsiella pneumoniae.Front Cell Infect Microbiol. 2023 Dec 1;13:1278482. doi: 10.3389/fcimb.2023.1278482. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 38106471 Free PMC article.
-
Diagnostic value of metagenomic next-generation sequencing in sepsis and bloodstream infection.Front Cell Infect Microbiol. 2023 Feb 10;13:1117987. doi: 10.3389/fcimb.2023.1117987. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 36844396 Free PMC article.
-
Comprehensive evaluation of plasma microbial cell-free DNA sequencing for predicting bloodstream and local infections in clinical practice: a multicenter retrospective study.Front Cell Infect Microbiol. 2024 Jan 4;13:1256099. doi: 10.3389/fcimb.2023.1256099. eCollection 2023. Front Cell Infect Microbiol. 2024. PMID: 38362158 Free PMC article.
-
Clinical and diagnostic values of metagenomic next-generation sequencing for infection in hematology patients: a systematic review and meta-analysis.BMC Infect Dis. 2024 Feb 7;24(1):167. doi: 10.1186/s12879-024-09073-x. BMC Infect Dis. 2024. PMID: 38326763 Free PMC article.
-
State-of-the-Art Metagenomic Sequencing and Its Role in the Diagnosis of Periprosthetic Joint Infections.Infect Dis Clin North Am. 2024 Dec;38(4):813-825. doi: 10.1016/j.idc.2024.07.011. Epub 2024 Sep 13. Infect Dis Clin North Am. 2024. PMID: 39277504 Review.
Cited by
-
Intensive Care Unit-onset Bloodstream Infections Represent a Distinct Category of Hospital-onset Infections: A Multicentre, Retrospective Cohort Study. Queensland Critical Care Network (QCCRN).J Assoc Med Microbiol Infect Dis Can. 2024 Dec 19;9(4):229-238. doi: 10.3138/jammi-2024-0023. eCollection 2024 Dec. J Assoc Med Microbiol Infect Dis Can. 2024. PMID: 40672712 Free PMC article.
-
Rapid and precise identification of cervicothoracic necrotizing fasciitis caused by Prevotella and Streptococcus constellatus by using Nanopore sequencing technology: a case report.Front Med (Lausanne). 2024 Oct 21;11:1447703. doi: 10.3389/fmed.2024.1447703. eCollection 2024. Front Med (Lausanne). 2024. PMID: 39497848 Free PMC article.
-
Application of Metagenomic Next-Generation Sequencing in HIV-Infected Patients with Bloodstream Infections.Infect Drug Resist. 2025 May 8;18:2389-2399. doi: 10.2147/IDR.S509665. eCollection 2025. Infect Drug Resist. 2025. PMID: 40357419 Free PMC article.
-
Diagnostic performance of metagenomic sequencing in patients with suspected infection: a large-scale retrospective study.Front Cell Infect Microbiol. 2024 Sep 6;14:1463081. doi: 10.3389/fcimb.2024.1463081. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39310785 Free PMC article.
-
Evaluation of Different Blood Culture Bottles for the Diagnosis of Bloodstream Infections in Patients with HIV.Infect Dis Ther. 2023 Nov;12(11):2611-2620. doi: 10.1007/s40121-023-00883-1. Epub 2023 Oct 23. Infect Dis Ther. 2023. PMID: 37870693 Free PMC article.
References
-
- Amar Y., Lagkouvardos I., Silva R. L., Ishola O. A., Foesel B. U., Kublik S., et al. . (2021). Pre-digest of unprotected DNA by benzonase improves the representation of living skin bacteria and efficiently depletes host DNA. Microbiome 9 (1), 123. doi: 10.1186/s40168-021-01067-0 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

