Immunological modifications following chemotherapy are associated with delayed recurrence of ovarian cancer
- PMID: 37435088
- PMCID: PMC10331425
- DOI: 10.3389/fimmu.2023.1204148
Immunological modifications following chemotherapy are associated with delayed recurrence of ovarian cancer
Abstract
Introduction: Ovarian cancer recurs in most High Grade Serous Ovarian Cancer (HGSOC) patients, including initial responders, after standard of care. To improve patient survival, we need to identify and understand the factors contributing to early or late recurrence and therapeutically target these mechanisms. We hypothesized that in HGSOC, the response to chemotherapy is associated with a specific gene expression signature determined by the tumor microenvironment. In this study, we sought to determine the differences in gene expression and the tumor immune microenvironment between patients who show early recurrence (within 6 months) compared to those who show late recurrence following chemotherapy.
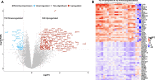
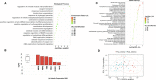
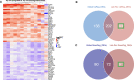
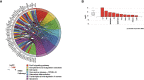
Methods: Paired tumor samples were obtained before and after Carboplatin and Taxol chemotherapy from 24 patients with HGSOC. Bioinformatic transcriptomic analysis was performed on the tumor samples to determine the gene expression signature associated with differences in recurrence pattern. Gene Ontology and Pathway analysis was performed using AdvaitaBio's iPathwayGuide software. Tumor immune cell fractions were imputed using CIBERSORTx. Results were compared between late recurrence and early recurrence patients, and between paired pre-chemotherapy and post-chemotherapy samples.
Results: There was no statistically significant difference between early recurrence or late recurrence ovarian tumors pre-chemotherapy. However, chemotherapy induced significant immunological changes in tumors from late recurrence patients but had no impact on tumors from early recurrence patients. The key immunological change induced by chemotherapy in late recurrence patients was the reversal of pro-tumor immune signature.
Discussion: We report for the first time, the association between immunological modifications in response to chemotherapy and the time of recurrence. Our findings provide novel opportunities to ultimately improve ovarian cancer patient survival.
Keywords: chemoresistance; cold tumors; hot tumors; immune response; ovarian cancer.
Copyright © 2023 Adzibolosu, Alvero, Ali-Fehmi, Gogoi, Corey, Tedja, Chehade, Gogoi, Morris, Anderson, Vitko, Lam, Craig, Draghici, Rutherford and Mor.
Conflict of interest statement
Author SD was employed by the company Advaita Corporation. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures









References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

