Screening of key genes related to ferroptosis and a molecular interaction network analysis in colorectal cancer using machine learning and bioinformatics
- PMID: 37435214
- PMCID: PMC10331738
- DOI: 10.21037/jgo-23-405
Screening of key genes related to ferroptosis and a molecular interaction network analysis in colorectal cancer using machine learning and bioinformatics
Abstract
Background: This study sought to identify the key genes and molecular interactions related to ferroptosis in colorectal cancer (CRC) using machine-learning and bioinformatics analyses.
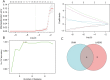
Methods: The Gene Expression Omnibus (National Institutes of Health, US) datasets for CRC were downloaded from the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/). The 291 ferroptosis genes were downloaded and screened from the FerrDb (http://www.zhounan.org/ferrdb) and GeneCards (https://www.genecards.org/) databases. The least absolute shrinkage and selection operator regression model and support vector machine model were constructed to identify the different ferroptosis-related hub genes. The immune infiltrates were identified and a survival curve analysis was conducted.
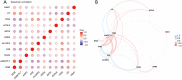
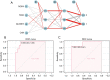
Results: We identified 11 ferroptosis-related differentially expressed genes (DEGs) from the COADREAD (Colon and Rectal Cancer) dataset. We found that angiopoietin-related protein 7 (ANGPTL7) gene expression was positively correlated to both the neuroglobin (NGB) (r=0.678) and ceruloplasmin (CP) (r=0.454) genes but was negatively correlated with transferrin receptor 2 (TFR2) expression (r=-0.426). In addition, TFR2 gene expression was positively correlated with the arachidonate lipoxygenase 3 (ALOXE3) (r=0.452) and carbonic anhydrase 9 (CA9) (r=0.411) genes. A total of 4 hub genes were identified by the machine-learning analysis [i.e., NADPH oxidase 4 (NOX4), TFR2, ALOXE3, and CA9]. The expression of the NOX4 gene was significantly positively correlated with neutrophil (r=0.543) and M0 macrophage (r=0.422) infiltration. In addition, a positive correlation between ALOXE3 and activated natural-killer cells (r=0.356) was found. Conversely, the NOX4, TFR2, and CA9 genes were negatively correlated with the resting mast cells. A strong negative correlation was observed between NOX4 and CD160 antigen (CD160) expression; however, a significant positive correlation was observed between NOX4 and transforming growth factor beta receptor 1 (TGFBR1) expression (r=0.397). The patients had a more favorable prognosis when the NOX4 expression levels were relatively low.
Conclusions: Our study identified 4 ferroptosis-related DEGs in CRC (i.e., NOX4, TFR2, ALOXE3, and CA9), and further validated their relationship with immune cell infiltration and the associated immune checkpoints. Our findings confirm the influence of the immune microenvironment on CRC. Low NOX4 levels were more favorable to patient outcomes. Our findings may facilitate future clinical diagnoses and outcome assessments of CRC.
Keywords: Ferroptosis-related genes; colorectal cancer (CRC); machine learning and bioinformatics; molecular interaction network.
2023 Journal of Gastrointestinal Oncology. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-405/coif). The authors have no conflicts of interest to declare.
Figures






Similar articles
-
Identification of a prognostic signature based on five ferroptosis-related genes for diffuse large B-cell lymphoma.Cancer Biomark. 2024;40(1):125-139. doi: 10.3233/CBM-230325. Cancer Biomark. 2024. PMID: 38517778 Free PMC article.
-
Identifification and validation of ferroptosis signatures and immune infifiltration characteristics associated with intervertebral disc degeneration.Front Genet. 2023 Feb 22;14:1133615. doi: 10.3389/fgene.2023.1133615. eCollection 2023. Front Genet. 2023. PMID: 36911415 Free PMC article.
-
Identification of Ferroptosis-related potential biomarkers and immunocyte characteristics in Chronic Thromboembolic Pulmonary Hypertension via bioinformatics analysis.BMC Cardiovasc Disord. 2023 Oct 11;23(1):504. doi: 10.1186/s12872-023-03511-5. BMC Cardiovasc Disord. 2023. PMID: 37821869 Free PMC article.
-
NOX4 has the potential to be a biomarker associated with colon cancer ferroptosis and immune infiltration based on bioinformatics analysis.Front Oncol. 2022 Sep 28;12:968043. doi: 10.3389/fonc.2022.968043. eCollection 2022. Front Oncol. 2022. PMID: 36249057 Free PMC article.
-
Ferroptosis-related NFE2L2 and NOX4 Genes are Potential Risk Prognostic Biomarkers and Correlated with Immunogenic Features in Glioma.Cell Biochem Biophys. 2023 Mar;81(1):7-17. doi: 10.1007/s12013-022-01124-x. Epub 2023 Jan 11. Cell Biochem Biophys. 2023. PMID: 36627482 Free PMC article. Review.
Cited by
-
Nutritional Modulation of Hepcidin in the Treatment of Various Anemic States.Nutrients. 2023 Dec 12;15(24):5081. doi: 10.3390/nu15245081. Nutrients. 2023. PMID: 38140340 Free PMC article. Review.
-
Co-vulnerabilities of inhibiting carbonic anhydrase IX in ferroptosis-mediated tumor cell death.Front Mol Biosci. 2023 Nov 30;10:1327310. doi: 10.3389/fmolb.2023.1327310. eCollection 2023. Front Mol Biosci. 2023. PMID: 38099193 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous
