Function of hsa_circ_0006646 as a competing endogenous RNA to promote progression in gastric cancer by regulating the miR-665-HMGB1 axis
- PMID: 37435216
- PMCID: PMC10331750
- DOI: 10.21037/jgo-23-240
Function of hsa_circ_0006646 as a competing endogenous RNA to promote progression in gastric cancer by regulating the miR-665-HMGB1 axis
Abstract
Background: Mounting evidences indicate that circular RNAs (circRNAs) are a novel class of non-coding RNAs and play vital roles in the tumorigenesis and aggressiveness including gastric cancer (GC). Nevertheless, the precise functions and underlying mechanisms of circRNAs in GC remain largely unknown.
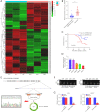
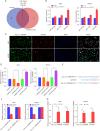
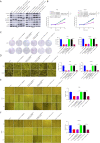
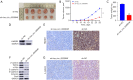
Methods: The Gene Expression Omnibus (GEO) data set GSE163416 was analyzed to screen the key circRNAs in GC. hsa_circ_0006646 was chosen for further study. GC tissues and matched adjacent normal gastric mucosal epithelial tissues were obtained from the Fourth Hospital of Hebei Medical University. The expressions of hsa_circ_0006646 was detected using quantitative real-time polymerase chain reaction (qRT-PCR). hsa_circ_0006646 was knocked down to identify its effects on GC cells. Bioinformatics algorithms were analyzed to predict the microRNA (miRNAs) potentially sponged by hsa_circ_0006646 and its target genes. Fluorescence in situ hybridization (FISH) was conducted to determine the subcellular location of hsa_circ_0006646 and the predicted miRNA. Then, qRT-PCR, luciferase reporter assay, radioimmunoprecipitation assay, Western blotting, and miRNA rescue experiments were used to confirm the hsa_circ_0006646-related regulatory axis in GC. Cell Counting Kit-8 (CCK-8), colony formation, wound healing, and Transwell experiments were performed to determine the effect of the hsa_circ_0006646-related regulatory axis on GC cells' malignant behaviors in vitro. The xenograft tumor mouse model was established to evaluate the effect of hsa_circ_0006646 in vivo.
Results: hsa_circ_0006646 exhibited a high expression in GC tissues as compared to corresponding adjacent normal gastric mucosal epithelial tissues and its high expression was positively correlated with TNM stage, lymph node invasion and poor prognosis (P<0.05). Knockdown of hsa_circ_0006646 suppressed the proliferation, colony formation, migration, and invasion in GC cells (all P<0.05). hsa_circ_0006646 upregulated high mobility group box 1 (HMGB1) by sponging miR-665 in GC cells (P<0.05). The hsa_circ_0006646-miR-665-HMGB1 axis promoted malignant behaviors and epithelial-mesenchymal transition (EMT) in GC cells by activating the Wnt/β-catenin pathway (P<0.05). The existence of hsa_circ_0006646-miR-665-HMGB1 axis was confirmed in GC specimens (P<0.05). Consequently, down-regulated hsa_circ_0006646 inhibited the progression and EMT of GC cells in vivo (P<0.05).
Conclusions: For the first time, we demonstrated that hsa_circ_0006646-miR-665-HMGB1 axis exerted its tumor-promoting effects in GC, which suggested that hsa_circ_0006646 could be potentially targeted for GC treatment.
Keywords: Gastric cancer (GC); epithelial-mesenchymal transition (EMT); high mobility group box 1 (HMGB1); hsa_circ_0006646; miR-665.
2023 Journal of Gastrointestinal Oncology. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-240/coif). The authors have no conflicts of interest to declare.
Figures


Similar articles
-
Circular RNA hsa_circ_0004872 inhibits gastric cancer progression via the miR-224/Smad4/ADAR1 successive regulatory circuit.Mol Cancer. 2020 Nov 10;19(1):157. doi: 10.1186/s12943-020-01268-5. Mol Cancer. 2020. PMID: 33172486 Free PMC article.
-
circ_0007385 served as competing endogenous RNA for miR-519d-3p to suppress malignant behaviors and cisplatin resistance of non-small cell lung cancer cells.Thorac Cancer. 2020 Aug;11(8):2196-2208. doi: 10.1111/1759-7714.13527. Epub 2020 Jun 29. Thorac Cancer. 2020. PMID: 32602212 Free PMC article.
-
Hsa_circ_0005230 is up-regulated and promotes gastric cancer cell invasion and migration via regulating the miR-1299/RHOT1 axis.Bioengineered. 2022 Mar;13(3):5046-5063. doi: 10.1080/21655979.2022.2036514. Bioengineered. 2022. PMID: 35170374 Free PMC article.
-
Circ-CSPP1 knockdown suppresses hepatocellular carcinoma progression through miR-493-5p releasing-mediated HMGB1 downregulation.Cell Signal. 2021 Oct;86:110065. doi: 10.1016/j.cellsig.2021.110065. Epub 2021 Jun 26. Cell Signal. 2021. PMID: 34182091 Review.
-
The potential of circular RNAs as biomarkers and therapeutic targets for gastric cancer: A comprehensive review.J Adv Res. 2024 Nov 29:S2090-1232(24)00551-4. doi: 10.1016/j.jare.2024.11.032. Online ahead of print. J Adv Res. 2024. PMID: 39617262 Review.
Cited by
-
Classical biomarkers and non-coding RNAs associated with diagnosis and treatment in gastric cancer.Oncol Res. 2025 Apr 18;33(5):1069-1089. doi: 10.32604/or.2025.063005. eCollection 2025. Oncol Res. 2025. PMID: 40296904 Free PMC article. Review.
-
Circular RNAs: the next level of gene regulation.Am J Transl Res. 2023 Oct 15;15(10):6122-6135. eCollection 2023. Am J Transl Res. 2023. PMID: 37969203 Free PMC article.
-
Unraveling the crosstalk: circRNAs and the wnt signaling pathway in cancers of the digestive system.Noncoding RNA Res. 2024 Apr 3;9(3):853-864. doi: 10.1016/j.ncrna.2024.03.004. eCollection 2024 Sep. Noncoding RNA Res. 2024. PMID: 38586314 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous
