The differential plasma and ruminal metabolic pathways and ruminal bacterial taxa associated with divergent residual body weight gain phenotype in crossbred beef steers
- PMID: 37435477
- PMCID: PMC10332501
- DOI: 10.1093/tas/txad054
The differential plasma and ruminal metabolic pathways and ruminal bacterial taxa associated with divergent residual body weight gain phenotype in crossbred beef steers
Abstract
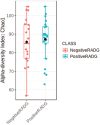
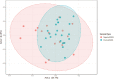
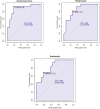
We applied ruminal and plasma metabolomics and ruminal 16S rRNA gene sequencing to determine the metabolic pathways and ruminal bacterial taxa associated with divergent residual body weight gain phenotype in crossbred beef steers. A group of 108 crossbred growing beef steers (average BW = 282.87 ± 30 kg) were fed a forage-based diet for a period of 56 d in a confinement dry lot equipped with GrowSafe intake nodes to determine their residual body weight gain (RADG) phenotype. After RADG identification, blood and rumen fluid samples were collected from beef steers with the highest RADG (most efficient; n = 16; 0.76 kg/d) and lowest RADG (least efficient; n = 16; -0.65 kg/d). Quantitative untargeted metabolome analysis of the plasma and rumen fluid samples were conducted using chemical isotope labelling/liquid chromatography-mass spectrometry. Differentially abundant metabolites in each of the plasma and rumen fluid samples between the two groups of beef steers were determined using a false discovery rate (FDR)-adjusted P-values ≤ 0.05 and area under the curve (AUC) > 0.80. Rumen and plasma metabolic pathways that were differentially enriched or depleted (P ≤ 0.05) in beef steers with positive RADG compared to those with negative RADG were determined by the quantitative pathway enrichment analysis. A total of 1,629 metabolites were detected and identified in the plasma of the beef steers; eight metabolites including alanyl-phenylalanine, 8-hydroxyguanosine, and slaframine were differentially abundant (FDR ≤ 0.05; AUC > 0.80) in beef steers with divergent RADG; five metabolic pathways including steroid hormone biosynthesis, thiamine metabolism, propanoate metabolism, pentose phosphate pathway, and butanoate metabolism were enriched (P ≤ 0.05) in beef steers with positive RADG, relative to negative RADG steers. A total of 1,908 metabolites were detected and identified in the rumen of the beef steers; results of the pathway enrichment analysis of all the metabolites revealed no metabolic pathways in the rumen were altered (P > 0.05). The rumen fluid samples were also analyzed using 16S rRNA gene sequencing to assess the bacterial community composition. We compared the rumen bacterial community composition at the genus level using a linear discriminant analysis effect size (LEfSe) to identify the differentially abundant taxa between the two groups of beef steers. The LEfSe results showed greater relative abundance of Bacteroidetes_vadinHA17 and Anaerovibrio in steers with positive RADG compared to the negative RADG group, while steers in the negative RADG group had greater relative abundance of Candidatus_Amoebophilus, Clostridium_sensu_stricto_1, Pseudomonas, Empedobacter, Enterobacter, and Klebsiella compared to the positive RADG group. Our results demonstrate that beef steers with positive or negative RADG exhibit differences in plasma metabolic profiles and some ruminal bacterial taxa which probably explain their divergent feed efficiency phenotypes.
Keywords: feed efficiency; metabolomics; rumen microbiome; steroid hormone.
© The Author(s) 2023. Published by Oxford University Press on behalf of the American Society of Animal Science.
Conflict of interest statement
None declared.
Figures


Similar articles
-
Effects of dietary supplementation of a blend of Saccharomyces cerevisiae, multiple live probiotic bacteria, and their fermentation products on performance, health, and rumen bacterial community of newly weaned beef steers during a 56-d receiving period.Transl Anim Sci. 2023 Dec 21;8:txad143. doi: 10.1093/tas/txad143. eCollection 2024. Transl Anim Sci. 2023. PMID: 38221963 Free PMC article.
-
Plasma proteomic analysis reveals key pathways associated with divergent residual body weight gain phenotype in beef steers.Front Vet Sci. 2024 Jul 22;11:1415594. doi: 10.3389/fvets.2024.1415594. eCollection 2024. Front Vet Sci. 2024. PMID: 39104547 Free PMC article.
-
Characterization of rumen microbiome and immune genes expression of crossbred beef steers with divergent residual feed intake phenotypes.BMC Genomics. 2024 Mar 5;25(1):245. doi: 10.1186/s12864-024-10150-3. BMC Genomics. 2024. PMID: 38443809 Free PMC article.
-
Rumen and plasma metabolomics profiling by UHPLC-QTOF/MS revealed metabolic alterations associated with a high-corn diet in beef steers.PLoS One. 2018 Nov 28;13(11):e0208031. doi: 10.1371/journal.pone.0208031. eCollection 2018. PLoS One. 2018. PMID: 30485366 Free PMC article.
-
Identification of Key Pathways Associated With Residual Feed Intake of Beef Cattle Based on Whole Blood Transcriptome Data Analyzed Using Gene Set Enrichment Analysis.Front Vet Sci. 2022 Apr 18;9:848027. doi: 10.3389/fvets.2022.848027. eCollection 2022. Front Vet Sci. 2022. PMID: 35518641 Free PMC article.
Cited by
-
Unraveling Ruminant Feed Efficiency Through Metabolomics: A Systematic Review.Metabolites. 2024 Dec 3;14(12):675. doi: 10.3390/metabo14120675. Metabolites. 2024. PMID: 39728456 Free PMC article. Review.
-
Effects of dietary supplementation of a blend of Saccharomyces cerevisiae, multiple live probiotic bacteria, and their fermentation products on performance, health, and rumen bacterial community of newly weaned beef steers during a 56-d receiving period.Transl Anim Sci. 2023 Dec 21;8:txad143. doi: 10.1093/tas/txad143. eCollection 2024. Transl Anim Sci. 2023. PMID: 38221963 Free PMC article.
-
Effect of Dietary Addition of Lentinus edodes on Rumen Flora, Lactation, and Health of Dairy Goats.Animals (Basel). 2025 Feb 26;15(5):676. doi: 10.3390/ani15050676. Animals (Basel). 2025. PMID: 40075961 Free PMC article.
References
-
- Alfarouk, K. O., Ahmed S. B. M., Elliott R. L., Benoit A., Alqahtani S. S., Ibrahim M. E., Bashir A. H. H., Alhoufie S. T. S., Elhassan G. O., et al.. 2020. The pentose phosphate pathway dynamics in cancer and its dependency on intracellular pH. Met. 10:285. doi:10.3390/metabo10070285. - DOI - PMC - PubMed
-
- Bailey, J. N. C., Yaspan B. L., Pasquale L. R., Hauser M. A., Kang J. H., Loomis S. J., Brilliant M., Budenz D. L., Christen W. G., Fingert J., et al.. 2014. Hypothesis-independent pathway analysis implicates GABA and Acetyl-CoA metabolism in primary open-angle glaucoma and normal-pressure glaucoma. Hum. Gen. 133:1319–1330. doi: 10.1007/s00439-014-1468-7. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
