DELLA proteins positively regulate seed size in Arabidopsis
- PMID: 37435751
- PMCID: PMC10445750
- DOI: 10.1242/dev.201853
DELLA proteins positively regulate seed size in Arabidopsis
Abstract
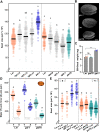
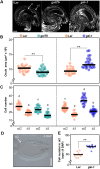
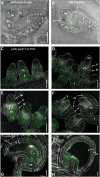
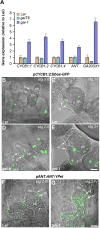
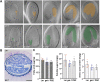
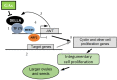
Human and animal nutrition is mainly based on seeds. Seed size is a key factor affecting seed yield and has thus been one of the primary objectives of plant breeders since the domestication of crop plants. Seed size is coordinately regulated by signals of maternal and zygotic tissues that control the growth of the seed coat, endosperm and embryo. Here, we provide previously unreported evidence for the role of DELLA proteins, key repressors of gibberellin responses, in the maternal control of seed size. The gain-of-function della mutant gai-1 produces larger seeds as a result of an increase in the cell number in ovule integuments. This leads to an increase in ovule size and, in turn, to an increase in seed size. Moreover, DELLA activity promotes increased seed size by inducing the transcriptional activation of AINTEGUMENTA, a genetic factor that controls cell proliferation and organ growth, in the ovule integuments of gai-1. Overall, our results indicate that DELLA proteins are involved in the control of seed size and suggest that modulation of the DELLA-dependent pathway could be used to improve crop yield.
Keywords: Arabidopsis; DELLA; Gibberellin; Ovule; Seed.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures
References
-
- Barro-Trastoy, D., Carrera, E., Baños, J., Palau-Rodriguez, J., Ruiz-Rivero, O., Tornero, P., Alonso, J. M., Lopez-Diaz, I., Gomez, M. D. and Perez-Amador, M. A. (2020). Regulation of ovule initiation by gibberellins and brassinosteroids in tomato and Arabidopsis: two plant species, two molecular mechanisms. Plant J. 102, 1026-1041. 10.1111/tpj.14684 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

