Analysis of Bacterial Phosphorylcholine-Related Genes Reveals an Association between Type-Specific Biosynthesis Pathways and Biomolecules Targeted for Phosphorylcholine Modification
- PMID: 37436144
- PMCID: PMC10434233
- DOI: 10.1128/spectrum.01583-23
Analysis of Bacterial Phosphorylcholine-Related Genes Reveals an Association between Type-Specific Biosynthesis Pathways and Biomolecules Targeted for Phosphorylcholine Modification
Abstract
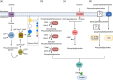
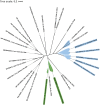
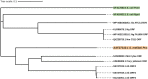
Many bacterial surface proteins and carbohydrates are modified with phosphorylcholine (ChoP), which contributes to host mimicry and can also promote colonization and survival in the host. However, the ChoP biosynthetic pathways that are used in bacterial species that express ChoP have not been systematically studied. For example, the well-studied Lic-1 pathway is absent in some ChoP-expressing bacteria, such as Neisseria meningitidis and Neisseria gonorrhoeae. This raises a question as to the origin of the ChoP used for macromolecule biosynthesis in these species. In the current study, we used in silico analyses to identify the potential pathways involved in ChoP biosynthesis in genomes of the 26 bacterial species reported to express a ChoP-modified biomolecule. We used the four known ChoP biosynthetic pathways and a ChoP transferase as search terms to probe for their presence in these genomes. We found that the Lic-1 pathway is primarily associated with organisms producing ChoP-modified carbohydrates, such as lipooligosaccharide. Pilin phosphorylcholine transferase A (PptA) homologs were detected in all bacteria that express ChoP-modified proteins. Additionally, ChoP biosynthesis pathways, such as phospholipid N-methyltransferase (PmtA), phosphatidylcholine synthase (Pcs), or the acylation-dependent phosphatidylcholine biosynthesis pathway, which generate phosphatidylcholine, were also identified in species that produce ChoP-modified proteins. Thus, a major finding of this study is the association of a particular ChoP biosynthetic pathway with a cognate, target ChoP-modified surface factor; i.e., protein versus carbohydrate. This survey failed to identify a known biosynthetic pathway for some species that express ChoP, indicating that a novel ChoP biosynthetic pathway(s) may remain to be identified. IMPORTANCE The modification of bacterial surface virulence factors with phosphorylcholine (ChoP) plays an important role in bacterial virulence and pathogenesis. However, the ChoP biosynthetic pathways in bacteria have not been fully understood. In this study, we used in silico analysis to identify potential ChoP biosynthetic pathways in bacteria that express ChoP-modified biomolecules and found the association between a specific ChoP biosynthesis pathway and the cognate target ChoP-modified surface factor.
Keywords: ChoP; bacterial virulence; phosphatidylcholine; phosphoethanolamine; phosphorylcholine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





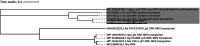
Similar articles
-
Identification and characterization of pptA: a gene involved in the phase-variable expression of phosphorylcholine on pili of Neisseria meningitidis.Infect Immun. 2003 Dec;71(12):6892-8. doi: 10.1128/IAI.71.12.6892-6898.2003. Infect Immun. 2003. PMID: 14638777 Free PMC article.
-
Substrate recognition of a structure motif for phosphorylcholine post-translational modification in Neisseria meningitidis.Biochem Biophys Res Commun. 2013 Feb 22;431(4):808-14. doi: 10.1016/j.bbrc.2012.12.088. Epub 2012 Dec 27. Biochem Biophys Res Commun. 2013. PMID: 23274496
-
The biosynthesis and role of phosphorylcholine in pathogenic and nonpathogenic bacteria.Trends Microbiol. 2023 Jul;31(7):692-706. doi: 10.1016/j.tim.2023.01.006. Epub 2023 Feb 28. Trends Microbiol. 2023. PMID: 36863982 Free PMC article. Review.
-
Genetic characterization of pilin glycosylation and phase variation in Neisseria meningitidis.Mol Microbiol. 2003 Aug;49(3):833-47. doi: 10.1046/j.1365-2958.2003.03602.x. Mol Microbiol. 2003. PMID: 12864863
-
Microbial modulation of host immunity with the small molecule phosphorylcholine.Infect Immun. 2013 Feb;81(2):392-401. doi: 10.1128/IAI.01168-12. Epub 2012 Dec 10. Infect Immun. 2013. PMID: 23230294 Free PMC article. Review.
Cited by
-
Substrate flexibility of Mycoplasma fermentans mf1 phosphorylcholine transferase.Glycoconj J. 2025 Apr;42(2):87-96. doi: 10.1007/s10719-025-10181-2. Epub 2025 Mar 22. Glycoconj J. 2025. PMID: 40119989 Free PMC article.
-
Broad specificity of monoclonal IgA (TEPC15-IgA) for enteric bacteria via phosphorylcholine-mediated interaction.J Vet Med Sci. 2024 Jul 2;86(7):801-808. doi: 10.1292/jvms.23-0441. Epub 2024 Jun 4. J Vet Med Sci. 2024. PMID: 38839348 Free PMC article.
References
-
- Fischer W, Behr T, Hartmann R, Peter-Katalinić J, Egge H. 1993. Teichoic acid and lipoteichoic acid of Streptococcus pneumoniae possess identical chain structures. A reinvestigation of teichoid acid (C polysaccharide). Eur J Biochem 215:851–857. doi:10.1111/j.1432-1033.1993.tb18102.x. - DOI - PubMed
-
- Kulakowska M, Brisson J-R, Griffith DW, Young NM, Jennings HJ. 1993. High-resolution NMR spectroscopic analysis of the C-polysaccharide of Streptococcus pneumoniae. Can J Chem 71:644–648. doi:10.1139/v93-086. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

