Selective Pressure by Rifampicin Modulates Mutation Rates and Evolutionary Trajectories of Mycobacterial Genomes
- PMID: 37436169
- PMCID: PMC10433840
- DOI: 10.1128/spectrum.01017-23
Selective Pressure by Rifampicin Modulates Mutation Rates and Evolutionary Trajectories of Mycobacterial Genomes
Abstract
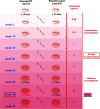
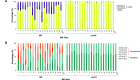
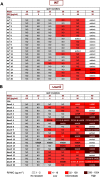
Resistance to the frontline antibiotic rifampicin constitutes a challenge to the treatment and control of tuberculosis. Here, we analyzed the mutational landscape of Mycobacterium smegmatis during long-term evolution with increasing concentrations of rifampicin, using a mutation accumulation assay combined with whole-genome sequencing. Antibiotic treatment enhanced the acquisition of mutations, doubling the genome-wide mutation rate of the wild-type cells. While antibiotic exposure led to extinction of almost all wild-type lines, the hypermutable phenotype of the ΔnucS mutant strain (noncanonical mismatch repair deficient) provided an efficient response to the antibiotic, leading to high rates of survival. This adaptative advantage resulted in the emergence of higher levels of rifampicin resistance, an accelerated acquisition of drug resistance mutations in rpoB (β RNA polymerase), and a wider diversity of evolutionary pathways that led to drug resistance. Finally, this approach revealed a subset of adaptive genes under positive selection with rifampicin that could be associated with the development of antibiotic resistance. IMPORTANCE Rifampicin is the most important first-line antibiotic against mycobacterial infections, including tuberculosis, one of the top causes of death worldwide. Acquisition of rifampicin resistance constitutes a major global public health problem that makes the control of the disease challenging. Here, we performed an experimental evolution assay under antibiotic selection to analyze the response and adaptation of mycobacteria, leading to the acquisition of rifampicin resistance. This approach explored the total number of mutations that arose in the mycobacterial genomes under long-term rifampicin exposure, using whole-genome sequencing. Our results revealed the effect of rifampicin at a genomic level, identifying different mechanisms and multiple pathways leading to rifampicin resistance in mycobacteria. Moreover, this study detected that an increase in the rate of mutations led to enhanced levels of drug resistance and survival. In summary, all of these results could be useful to understand and prevent the emergence of drug-resistant isolates in mycobacterial infections.
Keywords: DNA repair; Mycobacterium; Mycobacterium smegmatis; antibiotic resistance; drug resistance evolution; evolution; experimental evolution; mutation; mutation accumulation; rifampicin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Noncanonical Mismatch Repair Protein NucS Modulates the Emergence of Antibiotic Resistance in Mycobacterium abscessus.Microbiol Spectr. 2022 Dec 21;10(6):e0222822. doi: 10.1128/spectrum.02228-22. Epub 2022 Oct 11. Microbiol Spectr. 2022. PMID: 36219122 Free PMC article.
-
Evolution of Extensively Drug-Resistant Tuberculosis over Four Decades: Whole Genome Sequencing and Dating Analysis of Mycobacterium tuberculosis Isolates from KwaZulu-Natal.PLoS Med. 2015 Sep 29;12(9):e1001880. doi: 10.1371/journal.pmed.1001880. eCollection 2015 Sep. PLoS Med. 2015. PMID: 26418737 Free PMC article.
-
Absence of hybridization with the wild-type and mutant rpoB probes in the Genotype MTBDRplus assay detects 'disputed' rifampicin mutations.Clin Microbiol Infect. 2018 Jul;24(7):781.e1-781.e3. doi: 10.1016/j.cmi.2017.11.021. Epub 2017 Dec 5. Clin Microbiol Infect. 2018. PMID: 29217277 Free PMC article.
-
Consensus numbering system for the rifampicin resistance-associated rpoB gene mutations in pathogenic mycobacteria.Clin Microbiol Infect. 2017 Mar;23(3):167-172. doi: 10.1016/j.cmi.2016.09.006. Epub 2016 Sep 21. Clin Microbiol Infect. 2017. PMID: 27664776 Review.
-
Progress on the non-canonical mismatch repair in Mycobacterium and its role in antibiotic resistance.Yi Chuan. 2023 Nov 20;45(11):1018-1027. doi: 10.16288/j.yczz.23-236. Yi Chuan. 2023. PMID: 38764267 Review.
Cited by
-
SNPs in genes related to the repair of damage to DNA in clinical isolates of M. tuberculosis: A transversal and longitudinal approach.PLoS One. 2024 Jun 25;19(6):e0295464. doi: 10.1371/journal.pone.0295464. eCollection 2024. PLoS One. 2024. PMID: 38917091 Free PMC article.
-
Dinucleotide codon substitutions as a signature of diversifying selection in Mycobacterium tuberculosis.Front Mol Biosci. 2025 Jul 2;12:1511372. doi: 10.3389/fmolb.2025.1511372. eCollection 2025. Front Mol Biosci. 2025. PMID: 40671870 Free PMC article.
References
-
- O’Neill J. 2014. Antimicrobial resistance: tackling a crisis for the health and wealth of nations. Review on antimicrobial resistance. https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Ta....
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

