Misregulation of cell cycle-dependent methylation of budding yeast CENP-A contributes to chromosomal instability
- PMID: 37436802
- PMCID: PMC10551700
- DOI: 10.1091/mbc.E23-03-0108
Misregulation of cell cycle-dependent methylation of budding yeast CENP-A contributes to chromosomal instability
Abstract
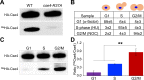
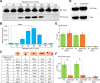
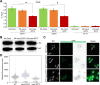
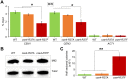
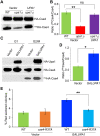
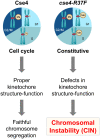
Centromere (CEN) identity is specified epigenetically by specialized nucleosomes containing evolutionarily conserved CEN-specific histone H3 variant CENP-A (Cse4 in Saccharomyces cerevisiae, CENP-A in humans), which is essential for faithful chromosome segregation. However, the epigenetic mechanisms that regulate Cse4 function have not been fully defined. In this study, we show that cell cycle-dependent methylation of Cse4-R37 regulates kinetochore function and high-fidelity chromosome segregation. We generated a custom antibody that specifically recognizes methylated Cse4-R37 and showed that methylation of Cse4 is cell cycle regulated with maximum levels of methylated Cse4-R37 and its enrichment at the CEN chromatin occur in the mitotic cells. Methyl-mimic cse4-R37F mutant exhibits synthetic lethality with kinetochore mutants, reduced levels of CEN-associated kinetochore proteins and chromosome instability (CIN), suggesting that mimicking the methylation of Cse4-R37 throughout the cell cycle is detrimental to faithful chromosome segregation. Our results showed that SPOUT methyltransferase Upa1 contributes to methylation of Cse4-R37 and overexpression of UPA1 leads to CIN phenotype. In summary, our studies have defined a role for cell cycle-regulated methylation of Cse4 in high-fidelity chromosome segregation and highlight an important role of epigenetic modifications such as methylation of kinetochore proteins in preventing CIN, an important hallmark of human cancers.
Figures


References
-
- Anedchenko EA, Samel-Pommerencke A, Tran Nguyen TM, Shahnejat-Bushehri S, Popsel J, Lauster D, Herrmann A, Rappsilber J, Cuomo A, Bonaldi T, Ehrenhofer-Murray AE (2019). The kinetochore module Okp1(CENP-Q)/Ame1(CENP-U) is a reader for N-terminal modifications on the centromeric histone Cse4(CENP-A). EMBO J 38, e98991. - PMC - PubMed
-
- Angrand G, Quillevere A, Loaec N, Dinh VT, Le Senechal R, Chennoufi R, Duchambon P, Keruzore M, Martins RP, Teulade-Fichou MP, et al. (2022). Type I arginine methyltransferases are intervention points to unveil the oncogenic Epstein-Barr virus to the immune system. Nucleic Acids Res 50, 11799–11819. - PMC - PubMed
-
- Au WC, Zhang T, Mishra PK, Eisenstatt JR, Walker RL, Ocampo J, Dawson A, Warren J, Costanzo M, Baryshnikova A, et al. (2020). Skp, Cullin, F-box (SCF)-Met30 and SCF-Cdc4-mediated proteolysis of CENP-A prevents mislocalization of CENP-A for chromosomal stability in budding yeast. PLoS Genet 16, e1008597. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HG005853/HG/NHGRI NIH HHS/United States
- R01 GM032238/GM/NIGMS NIH HHS/United States
- R37 GM032238/GM/NIGMS NIH HHS/United States
- BB/R00868X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/T017716/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

