Meiosis II spindle disassembly requires two distinct pathways
- PMID: 37436806
- PMCID: PMC10551701
- DOI: 10.1091/mbc.E23-03-0096
Meiosis II spindle disassembly requires two distinct pathways
Abstract
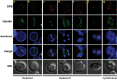
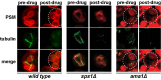
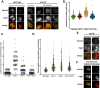
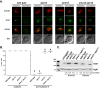
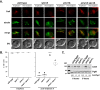
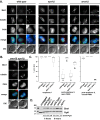
During exit from meiosis II, cells undergo several structural rearrangements, including disassembly of the meiosis II spindles and cytokinesis. Each of these changes is regulated to ensure that they occur at the proper time. Previous studies have demonstrated that both SPS1, which encodes a STE20-family GCKIII kinase, and AMA1, which encodes a meiosis-specific activator of the Anaphase Promoting Complex, are required for both meiosis II spindle disassembly and cytokinesis in the budding yeast Saccharomyces cerevisiae. We examine the relationship between meiosis II spindle disassembly and cytokinesis and find that the meiosis II spindle disassembly failure in sps1Δ and ama1∆ cells is not the cause of the cytokinesis defect. We also see that the spindle disassembly defects in sps1Δ and ama1∆ cells are phenotypically distinct. We examined known microtubule-associated proteins Ase1, Cin8, and Bim1, and found that AMA1 is required for the proper loss of Ase1 and Cin8 on meiosis II spindles while SPS1 is required for Bim1 loss in meiosis II. Taken together, these data indicate that SPS1 and AMA1 promote distinct aspects of meiosis II spindle disassembly, and that both pathways are required for the successful completion of meiosis.
Figures

References
-
- Amberg DC, Burke DJ, Strathern JN (2006). Inducing yeast cell synchrony: nocodazole arrest. CSH Protoc - PubMed
-
- Argüello-Miranda O, Zagoriy I, Mengoli V, Rojas J, Jonak K, Oz T, Graf P, Zachariae W (2017). Casein Kinase 1 coordinates cohesin cleavage, gametogenesis, and exit from M phase in meiosis II. Dev Cell 40, 37–52. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

