Improved HIV-1 neutralization breadth and potency of V2-apex antibodies by in silico design
- PMID: 37436900
- PMCID: PMC10528384
- DOI: 10.1016/j.celrep.2023.112711
Improved HIV-1 neutralization breadth and potency of V2-apex antibodies by in silico design
Abstract
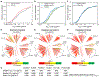
Broadly neutralizing antibodies (bNAbs) against HIV can reduce viral transmission in humans, but an effective therapeutic will require unusually high breadth and potency of neutralization. We employ the OSPREY computational protein design software to engineer variants of two apex-directed bNAbs, PGT145 and PG9RSH, resulting in increases in potency of over 100-fold against some viruses. The top designed variants improve neutralization breadth from 39% to 54% at clinically relevant concentrations (IC80 < 1 μg/mL) and improve median potency (IC80) by up to 4-fold over a cross-clade panel of 208 strains. To investigate the mechanisms of improvement, we determine cryoelectron microscopy structures of each variant in complex with the HIV envelope trimer. Surprisingly, we find the largest increases in breadth to be a result of optimizing side-chain interactions with highly variable epitope residues. These results provide insight into mechanisms of neutralization breadth and inform strategies for antibody design and improvement.
Keywords: CP: Immunology; CP: Microbiology; HIV; OSPREY; antibody design; antibody improvement; broadly neutralizing antibody; in silico design; protein design; provable algorithms; structural biology.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests B.R.D. and M.S.F. are founders of Ten63 Therapeutics. B.R.D., S.W., A.U.L., G.T.H., M.S.F., P.D.K., J.G., and N.A.D. are inventors on a patent application filed by Duke University.
Figures
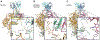

Similar articles
-
Structural development of the HIV-1 apex-directed PGT145-PGDM1400 antibody lineage.Cell Rep. 2025 Jan 28;44(1):115223. doi: 10.1016/j.celrep.2024.115223. Epub 2025 Jan 17. Cell Rep. 2025. PMID: 39826122 Free PMC article.
-
New Member of the V1V2-Directed CAP256-VRC26 Lineage That Shows Increased Breadth and Exceptional Potency.J Virol. 2015 Oct 14;90(1):76-91. doi: 10.1128/JVI.01791-15. Print 2016 Jan 1. J Virol. 2015. PMID: 26468542 Free PMC article.
-
Combinations of Single Chain Variable Fragments From HIV Broadly Neutralizing Antibodies Demonstrate High Potency and Breadth.Front Immunol. 2021 Sep 16;12:734110. doi: 10.3389/fimmu.2021.734110. eCollection 2021. Front Immunol. 2021. PMID: 34603312 Free PMC article.
-
Ontogeny-based immunogens for the induction of V2-directed HIV broadly neutralizing antibodies.Immunol Rev. 2017 Jan;275(1):217-229. doi: 10.1111/imr.12501. Immunol Rev. 2017. PMID: 28133797 Free PMC article. Review.
-
Antibody class-switching as a strategy to improve HIV-1 neutralization.Trends Mol Med. 2022 Nov;28(11):979-988. doi: 10.1016/j.molmed.2022.08.010. Epub 2022 Sep 15. Trends Mol Med. 2022. PMID: 36117072 Free PMC article. Review.
Cited by
-
Structural development of the HIV-1 apex-directed PGT145-PGDM1400 antibody lineage.Cell Rep. 2025 Jan 28;44(1):115223. doi: 10.1016/j.celrep.2024.115223. Epub 2025 Jan 17. Cell Rep. 2025. PMID: 39826122 Free PMC article.
-
DexDesign: an OSPREY-based algorithm for designing de novo D-peptide inhibitors.Protein Eng Des Sel. 2024 Jan 29;37:gzae007. doi: 10.1093/protein/gzae007. Protein Eng Des Sel. 2024. PMID: 38757573 Free PMC article.
-
Protocol for Designing De Novo Noncanonical Peptide Binders in OSPREY.J Comput Biol. 2024 Oct;31(10):965-974. doi: 10.1089/cmb.2024.0669. Epub 2024 Oct 4. J Comput Biol. 2024. PMID: 39364612
-
Cryo-electron microscopy in the study of virus entry and infection.Front Mol Biosci. 2024 Jul 24;11:1429180. doi: 10.3389/fmolb.2024.1429180. eCollection 2024. Front Mol Biosci. 2024. PMID: 39114367 Free PMC article. Review.
-
Predicting Affinity Through Homology (PATH): Interpretable binding affinity prediction with persistent homology.PLoS Comput Biol. 2025 Jun 27;21(6):e1013216. doi: 10.1371/journal.pcbi.1013216. eCollection 2025 Jun. PLoS Comput Biol. 2025. PMID: 40577377 Free PMC article.
References
-
- Rudicell RS, Kwon YD, Ko SY, Pegu A, Louder MK, Georgiev IS,Wu X, Zhu J, Boyington JC, Chen X, et al. (2014). Enhanced Potency of a Broadly Neutralizing HIV-1 Antibody In Vitro Improves Protection against Lentiviral Infection In Vivo. J. Virol 88, 12669–12682. 10.1128/JVI.02213-14. - DOI - PMC - PubMed
-
- Julg B, Tartaglia LJ, Keele BF, Wagh K, Pegu A, Sok D, Abbink P, Schmidt SD, Wang K, Chen X, et al. (2017). Broadly neutralizing antibodies targeting the HIV-1 envelope V2 apex confer protection against a clade C SHIV challenge. Sci. Transl. Med 9, eaal1321. 10.1126/scitranslmed.aal1321. - DOI - PMC - PubMed
-
- Pegu A, Borate B, Huang Y, Pauthner MG, Hessell AJ, Julg B, Do-ria-Rose NA, Schmidt SD, Carpp LN, Cully MD, et al. (2019). A Meta-analysis of Passive Immunization Studies Shows that Serum-Neutralizing Antibody Titer Associates with Protection against SHIV Challenge. Cell Host Microbe 26, 336–346.e3. 10.1016/j.chom.2019.08.014. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

