GRL-142 binds to and impairs HIV-1 integrase nuclear localization signal and potently suppresses highly INSTI-resistant HIV-1 variants
- PMID: 37436982
- PMCID: PMC10337902
- DOI: 10.1126/sciadv.adg2955
GRL-142 binds to and impairs HIV-1 integrase nuclear localization signal and potently suppresses highly INSTI-resistant HIV-1 variants
Abstract
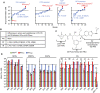
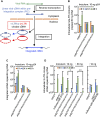
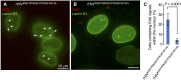
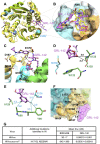
Nuclear localization signal (NLS) of HIV-1 integrase (IN) is implicated in nuclear import of HIV-1 preintegration complex (PIC). Here, we established a multiclass drug-resistant HIV-1 variant (HIVKGD) by consecutively exposing an HIV-1 variant to various antiretroviral agents including IN strand transfer inhibitors (INSTIs). HIVKGD was extremely susceptible to a previously reported HIV-1 protease inhibitor, GRL-142, with IC50 of 130 femtomolar. When cells were exposed to HIVKGD IN-containing recombinant HIV in the presence of GRL-142, significant decrease of unintegrated 2-LTR circular cDNA was observed, suggesting that nuclear import of PIC was severely compromised by GRL-142. X-ray crystallographic analyses revealed that GRL-142 interacts with NLS's putative sequence (DQAEHLK) and sterically blocks the nuclear transport of GRL-142-bound HIVKGD's PIC. Highly INSTI-resistant HIV-1 variants isolated from heavily INSTI-experienced patients proved to be susceptible to GRL-142, suggesting that NLS-targeting agents would serve as salvage therapy agents for highly INSTI-resistant variant-harboring individuals. The data should offer a new modality to block HIV-1 infectivity and replication and shed light on developing NLS inhibitors for AIDS therapy.
Figures


References
-
- J. L. Marcus, C. R. Chao, W. A. Leyden, L. Xu, C. P. Quesenberry Jr., D. B. Klein, W. J. Towner, M. A. Horberg, M. J. Silverberg, Narrowing the gap in life expectancy between HIV-infected and HIV-uninfected individuals with access to care. J. Acquir. Immune Defic. Syndr. 73, 39–46 (2016). - PMC - PubMed
-
- D. Armenia, D. Di Carlo, P. Flandre, Y. Bouba, V. Borghi, F. Forbici, A. Bertoli, C. Gori, L. Fabeni, W. Gennari, C. Pinnetti, A. Mondi, S. Cicalini, R. Gagliardini, A. Vergori, R. Bellagamba, V. Malagnino, F. Montella, M. Colafigli, A. Latini, R. Marocco, M. Licthner, M. Andreoni, C. Mussini, F. Ceccherini-Silberstein, A. Antinori, C. F. Perno, M. M. Santoro, HIV MDR is still a relevant issue despite its dramatic drop over the years. J. Antimicrob. Chemother. 75, 1301–1310 (2020). - PubMed
-
- R. M. Kagan, K. J. Dunn, G. P. Snell, R. E. Nettles, H. W. Kaufman, Trends in HIV-1 drug resistance mutations from a U.S. Reference Laboratory from 2006 to 2017. AIDS Res. Hum. Retroviruses 35, 698–709 (2019). - PubMed
-
- D. Grover, A. Copas, H. Green, S. G. Edwards, D. T. Dunn, C. Sabin, A. Phillips, E. Allen, D. Pillay; UK Collaborative Group on HIV Drug Resistance and UK Collaborative HIV Cohort Study (UK CHIC) , What is the risk of mortality following diagnosis of multidrug-resistant HIV-1? J. Antimicrob. Chemother. 61, 705–713 (2008). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

