Bacterial Pathogen Infection Triggers Magic Spot Nucleotide Signaling in Arabidopsis thaliana Chloroplasts through Specific RelA/SpoT Homologues
- PMID: 37437195
- PMCID: PMC10375528
- DOI: 10.1021/jacs.3c04445
Bacterial Pathogen Infection Triggers Magic Spot Nucleotide Signaling in Arabidopsis thaliana Chloroplasts through Specific RelA/SpoT Homologues
Abstract
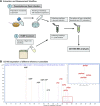
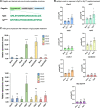
Magic spot nucleotides (p)ppGpp are important signaling molecules in bacteria and plants. In the latter, RelA-SpoT homologue (RSH) enzymes are responsible for (p)ppGpp turnover. Profiling of (p)ppGpp is more difficult in plants than in bacteria due to lower concentrations and more severe matrix effects. Here, we report that capillary electrophoresis mass spectrometry (CE-MS) can be deployed to study (p)ppGpp abundance and identity in Arabidopsis thaliana. This goal is achieved by combining a titanium dioxide extraction protocol and pre-spiking with chemically synthesized stable isotope-labeled internal reference compounds. The high sensitivity and separation efficiency of CE-MS enables monitoring of changes in (p)ppGpp levels in A. thaliana upon infection with the pathogen Pseudomonas syringae pv. tomato (PstDC3000). We observed a significant increase of ppGpp post infection that is also stimulated by the flagellin peptide flg22 only. This increase depends on functional flg22 receptor FLS2 and its interacting kinase BAK1 indicating that pathogen-associated molecular pattern (PAMP) receptor-mediated signaling controls ppGpp levels. Transcript analyses showed an upregulation of RSH2 upon flg22 treatment and both RSH2 and RSH3 after PstDC3000 infection. Arabidopsis mutants deficient in RSH2 and RSH3 activity display no ppGpp accumulation upon infection and flg22 treatment, supporting the involvement of these synthases in PAMP-triggered innate immune responses to pathogens within the chloroplast.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
Plastidial (p)ppGpp Synthesis by the Ca2+-Dependent RelA-SpoT Homolog Regulates the Adaptation of Chloroplast Gene Expression to Darkness in Arabidopsis.Plant Cell Physiol. 2021 Feb 4;61(12):2077-2086. doi: 10.1093/pcp/pcaa124. Plant Cell Physiol. 2021. PMID: 33089303
-
Arabidopsis RelA/SpoT homologs implicate (p)ppGpp in plant signaling.Proc Natl Acad Sci U S A. 2000 Mar 28;97(7):3747-52. doi: 10.1073/pnas.97.7.3747. Proc Natl Acad Sci U S A. 2000. PMID: 10725385 Free PMC article.
-
Expression profiling of four RelA/SpoT-like proteins, homologues of bacterial stringent factors, in Arabidopsis thaliana.Planta. 2008 Sep;228(4):553-62. doi: 10.1007/s00425-008-0758-5. Epub 2008 Jun 6. Planta. 2008. PMID: 18535838
-
Within and beyond the stringent response-RSH and (p)ppGpp in plants.Planta. 2017 Nov;246(5):817-842. doi: 10.1007/s00425-017-2780-y. Epub 2017 Sep 25. Planta. 2017. PMID: 28948393 Free PMC article. Review.
-
Signalling by the global regulatory molecule ppGpp in bacteria and chloroplasts of land plants.Plant Biol (Stuttg). 2011 Sep;13(5):699-709. doi: 10.1111/j.1438-8677.2011.00484.x. Epub 2011 May 31. Plant Biol (Stuttg). 2011. PMID: 21815973 Review.
Cited by
-
Exosome-like nanoparticles derived from fruits, vegetables, and herbs: innovative strategies of therapeutic and drug delivery.Theranostics. 2024 Aug 1;14(12):4598-4621. doi: 10.7150/thno.97096. eCollection 2024. Theranostics. 2024. PMID: 39239509 Free PMC article.
-
Effects of Light and Dark Conditions on the Transcriptome of Aging Cultures of Candidatus Puniceispirillum marinum IMCC1322.J Microbiol. 2024 Apr;62(4):297-314. doi: 10.1007/s12275-024-00125-0. Epub 2024 Apr 25. J Microbiol. 2024. PMID: 38662311
-
How to quantify magic spots - a brief overview of (p)ppGpp detection and quantitation methods.Front Mol Biosci. 2025 Mar 25;12:1574135. doi: 10.3389/fmolb.2025.1574135. eCollection 2025. Front Mol Biosci. 2025. PMID: 40201240 Free PMC article. Review.
-
Structural Insights into Broad-Range Polyphosphate Kinase 2-II Enzymes Applicable for Pyrimidine Nucleoside Diphosphate Synthesis.Chembiochem. 2025 Feb 3;26(5):e202400970. doi: 10.1002/cbic.202400970. Epub 2025 Feb 4. Chembiochem. 2025. PMID: 39846220 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

