Chemically induced reprogramming to reverse cellular aging
- PMID: 37437248
- PMCID: PMC10373966
- DOI: 10.18632/aging.204896
Chemically induced reprogramming to reverse cellular aging
Abstract
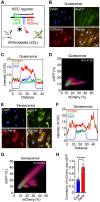
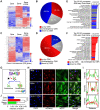
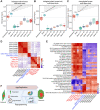
A hallmark of eukaryotic aging is a loss of epigenetic information, a process that can be reversed. We have previously shown that the ectopic induction of the Yamanaka factors OCT4, SOX2, and KLF4 (OSK) in mammals can restore youthful DNA methylation patterns, transcript profiles, and tissue function, without erasing cellular identity, a process that requires active DNA demethylation. To screen for molecules that reverse cellular aging and rejuvenate human cells without altering the genome, we developed high-throughput cell-based assays that distinguish young from old and senescent cells, including transcription-based aging clocks and a real-time nucleocytoplasmic compartmentalization (NCC) assay. We identify six chemical cocktails, which, in less than a week and without compromising cellular identity, restore a youthful genome-wide transcript profile and reverse transcriptomic age. Thus, rejuvenation by age reversal can be achieved, not only by genetic, but also chemical means.
Keywords: epigenetics; information theory of aging; rejuvenation medicine; reprogramming; small molecules.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

