Maintenance of neuronal identity in C. elegans and beyond: Lessons from transcription and chromatin factors
- PMID: 37438210
- PMCID: PMC10592372
- DOI: 10.1016/j.semcdb.2023.07.001
Maintenance of neuronal identity in C. elegans and beyond: Lessons from transcription and chromatin factors
Abstract
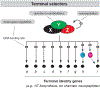
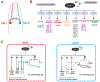
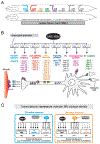
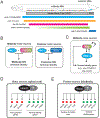
Neurons are remarkably long-lived, non-dividing cells that must maintain their functional features (e.g., electrical properties, chemical signaling) for extended periods of time - decades in humans. How neurons accomplish this incredible feat is poorly understood. Here, we review recent advances, primarily in the nematode C. elegans, that have enhanced our understanding of the molecular mechanisms that enable post-mitotic neurons to maintain their functionality across different life stages. We begin with "terminal selectors" - transcription factors necessary for the establishment and maintenance of neuronal identity. We highlight new findings on five terminal selectors (CHE-1 [Glass], UNC-3 [Collier/Ebf1-4], LIN-39 [Scr/Dfd/Hox4-5], UNC-86 [Acj6/Brn3a-c], AST-1 [Etv1/ER81]) from different transcription factor families (ZNF, COE, HOX, POU, ETS). We compare the functions of these factors in specific neuron types of C. elegans with the actions of their orthologs in other invertebrate (D. melanogaster) and vertebrate (M. musculus) systems, highlighting remarkable functional conservation. Finally, we reflect on recent findings implicating chromatin-modifying proteins, such as histone methyltransferases and Polycomb proteins, in the control of neuronal terminal identity. Altogether, these new studies on transcription factors and chromatin modifiers not only shed light on the fundamental problem of neuronal identity maintenance, but also outline mechanistic principles of gene regulation that may operate in other long-lived, post-mitotic cell types.
Keywords: C. elegans; Chromatin-modifying proteins; D. melanogaster; Maintenance; Mus musculus; Neuronal identity; Terminal selectors; Transcription factors.
Copyright © 2023 The Authors. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
Similar articles
-
Brn3/POU-IV-type POU homeobox genes-Paradigmatic regulators of neuronal identity across phylogeny.Wiley Interdiscip Rev Dev Biol. 2020 Jul;9(4):e374. doi: 10.1002/wdev.374. Epub 2020 Feb 3. Wiley Interdiscip Rev Dev Biol. 2020. PMID: 32012462 Review.
-
A terminal selector prevents a Hox transcriptional switch to safeguard motor neuron identity throughout life.Elife. 2020 Jan 3;9:e50065. doi: 10.7554/eLife.50065. Elife. 2020. PMID: 31902393 Free PMC article.
-
The LIM and POU homeobox genes ttx-3 and unc-86 act as terminal selectors in distinct cholinergic and serotonergic neuron types.Development. 2014 Jan;141(2):422-35. doi: 10.1242/dev.099721. Epub 2013 Dec 18. Development. 2014. PMID: 24353061 Free PMC article.
-
Maintenance of neurotransmitter identity by Hox proteins through a homeostatic mechanism.Nat Commun. 2022 Oct 15;13(1):6097. doi: 10.1038/s41467-022-33781-0. Nat Commun. 2022. PMID: 36243871 Free PMC article.
-
Terminal Selectors of Neuronal Identity.Curr Top Dev Biol. 2016;116:455-75. doi: 10.1016/bs.ctdb.2015.12.007. Epub 2016 Jan 14. Curr Top Dev Biol. 2016. PMID: 26970634 Review.
Cited by
-
Functions of nuclear factor Y in nervous system development, function and health.Neural Regen Res. 2025 Oct 1;20(10):2887-2894. doi: 10.4103/NRR.NRR-D-24-00684. Epub 2024 Oct 22. Neural Regen Res. 2025. PMID: 39610092 Free PMC article.
-
A molecular atlas of adult C. elegans motor neurons reveals ancient diversity delineated by conserved transcription factor codes.Cell Rep. 2024 Mar 26;43(3):113857. doi: 10.1016/j.celrep.2024.113857. Epub 2024 Feb 29. Cell Rep. 2024. PMID: 38421866 Free PMC article.
-
Interlocked transcription factor feedback loops maintain and restore touch sensation.bioRxiv [Preprint]. 2025 May 20:2025.05.15.654349. doi: 10.1101/2025.05.15.654349. bioRxiv. 2025. PMID: 40475441 Free PMC article. Preprint.
-
Decoding neuronal diversity: Mechanisms governing neural cell fate in Drosophila.Curr Opin Neurobiol. 2025 Aug;93:103061. doi: 10.1016/j.conb.2025.103061. Epub 2025 Jun 6. Curr Opin Neurobiol. 2025. PMID: 40482397 Review.
-
RLS-associated MEIS transcription factors control distinct processes in human neural stem cells.Sci Rep. 2024 Nov 22;14(1):28986. doi: 10.1038/s41598-024-80266-9. Sci Rep. 2024. PMID: 39578497 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

