Tolerance of repeated toxic injuries of murine livers is associated with steatosis and inflammation
- PMID: 37438332
- PMCID: PMC10338629
- DOI: 10.1038/s41419-023-05855-4
Tolerance of repeated toxic injuries of murine livers is associated with steatosis and inflammation
Abstract
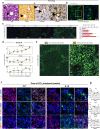
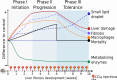
The human liver has a remarkable capacity to regenerate and thus compensate over decades for fibrosis caused by toxic chemicals, drugs, alcohol, or malnutrition. To date, no protective mechanisms have been identified that help the liver tolerate these repeated injuries. In this study, we revealed dysregulation of lipid metabolism and mild inflammation as protective mechanisms by studying longitudinal multi-omic measurements of liver fibrosis induced by repeated CCl4 injections in mice (n = 45). Based on comprehensive proteomics, transcriptomics, blood- and tissue-level profiling, we uncovered three phases of early disease development-initiation, progression, and tolerance. Using novel multi-omic network analysis, we identified multi-level mechanisms that are significantly dysregulated in the injury-tolerant response. Public data analysis shows that these profiles are altered in human liver diseases, including fibrosis and early cirrhosis stages. Our findings mark the beginning of the tolerance phase as the critical switching point in liver response to repetitive toxic doses. After fostering extracellular matrix accumulation as an acute response, we observe a deposition of tiny lipid droplets in hepatocytes only in the Tolerant phase. Our comprehensive study shows that lipid metabolism and mild inflammation may serve as biomarkers and are putative functional requirements to resist further disease progression.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

