Prospective validation of dermoscopy-based open-source artificial intelligence for melanoma diagnosis (PROVE-AI study)
- PMID: 37438476
- PMCID: PMC10338483
- DOI: 10.1038/s41746-023-00872-1
Prospective validation of dermoscopy-based open-source artificial intelligence for melanoma diagnosis (PROVE-AI study)
Abstract
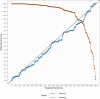
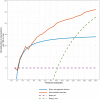
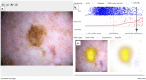
The use of artificial intelligence (AI) has the potential to improve the assessment of lesions suspicious of melanoma, but few clinical studies have been conducted. We validated the accuracy of an open-source, non-commercial AI algorithm for melanoma diagnosis and assessed its potential impact on dermatologist decision-making. We conducted a prospective, observational clinical study to assess the diagnostic accuracy of the AI algorithm (ADAE) in predicting melanoma from dermoscopy skin lesion images. The primary aim was to assess the reliability of ADAE's sensitivity at a predefined threshold of 95%. Patients who had consented for a skin biopsy to exclude melanoma were eligible. Dermatologists also estimated the probability of melanoma and indicated management choices before and after real-time exposure to ADAE scores. All lesions underwent biopsy. Four hundred thirty-five participants were enrolled and contributed 603 lesions (95 melanomas). Participants had a mean age of 59 years, 54% were female, and 96% were White individuals. At the predetermined 95% sensitivity threshold, ADAE had a sensitivity of 96.8% (95% CI: 91.1-98.9%) and specificity of 37.4% (95% CI: 33.3-41.7%). The dermatologists' ability to assess melanoma risk significantly improved after ADAE exposure (AUC 0.7798 vs. 0.8161, p = 0.042). Post-ADAE dermatologist decisions also had equivalent or higher net benefit compared to biopsying all lesions. We validated the accuracy of an open-source melanoma AI algorithm and showed its theoretical potential for improving dermatology experts' ability to evaluate lesions suspicious of melanoma. Larger randomized trials are needed to fully evaluate the potential of adopting this AI algorithm into clinical workflows.
© 2023. The Author(s).
Conflict of interest statement
V.R. is an expert advisor for Inhabit Brands. A.H. consults for Canfield Scientific Inc. and Janssen Research and Development, has ownership/equity interests in HCW Health LLC, SKIP Derm LLC, and has a fiduciary role/position and intellectual rights in SKIP Derm LLC. A.M. reports receiving honorarium from Canfield Scientific Inc. All other authors declare no competing interests.
Figures

References
-
- Daneshjou R, et al. Checklist for Evaluation of Image-Based Artificial Intelligence Reports in Dermatology: CLEAR Derm Consensus Guidelines From the International Skin Imaging Collaboration Artificial Intelligence Working Group. JAMA Dermatol. 2022;158:90–96. doi: 10.1001/jamadermatol.2021.4915. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

