Spatially resolved multiomics of human cardiac niches
- PMID: 37438528
- PMCID: PMC10371870
- DOI: 10.1038/s41586-023-06311-1
Spatially resolved multiomics of human cardiac niches
Erratum in
-
Author Correction: Spatially resolved multiomics of human cardiac niches.Nature. 2025 Apr;640(8058):E4. doi: 10.1038/s41586-025-08886-3. Nature. 2025. PMID: 40113895 Free PMC article. No abstract available.
Abstract
The function of a cell is defined by its intrinsic characteristics and its niche: the tissue microenvironment in which it dwells. Here we combine single-cell and spatial transcriptomics data to discover cellular niches within eight regions of the human heart. We map cells to microanatomical locations and integrate knowledge-based and unsupervised structural annotations. We also profile the cells of the human cardiac conduction system1. The results revealed their distinctive repertoire of ion channels, G-protein-coupled receptors (GPCRs) and regulatory networks, and implicated FOXP2 in the pacemaker phenotype. We show that the sinoatrial node is compartmentalized, with a core of pacemaker cells, fibroblasts and glial cells supporting glutamatergic signalling. Using a custom CellPhoneDB.org module, we identify trans-synaptic pacemaker cell interactions with glia. We introduce a druggable target prediction tool, drug2cell, which leverages single-cell profiles and drug-target interactions to provide mechanistic insights into the chronotropic effects of drugs, including GLP-1 analogues. In the epicardium, we show enrichment of both IgG+ and IgA+ plasma cells forming immune niches that may contribute to infection defence. Overall, we provide new clarity to cardiac electro-anatomy and immunology, and our suite of computational approaches can be applied to other tissues and organs.
© 2023. The Author(s).
Conflict of interest statement
In the past 3 years, S.A.T. has consulted or been a member of scientific advisory boards at Roche, Genentech, Biogen, GlaxoSmithKline, Qiagen and ForeSite Labs and is an equity holder of Transition Bio. The remaining authors declare no competing interests.
Figures







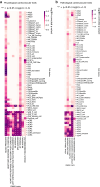



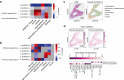





Comment in
-
Spatial transcriptomics of the human heart.Nat Biotechnol. 2023 Aug;41(8):1072. doi: 10.1038/s41587-023-01918-1. Nat Biotechnol. 2023. PMID: 37568021 No abstract available.
References
-
- van Eif, V. W. W., Devalla, H. D., Boink, G. J. J. & Christoffels, V. M. Transcriptional regulation of the cardiac conduction system. Nat. Rev. Cardiol.15, 617–630 (2018). - PubMed
-
- Miranda, A. M. A. et al. Single-cell transcriptomics for the assessment of cardiac disease. Nat. Rev. Cardiol.20, 289–308 (2023). - PubMed
-
- Asp, M. et al. A spatiotemporal organ-wide gene expression and cell atlas of the developing human heart. Cell179, 1647–1660.e19 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

