PLSCR1 is a cell-autonomous defence factor against SARS-CoV-2 infection
- PMID: 37438530
- PMCID: PMC10371867
- DOI: 10.1038/s41586-023-06322-y
PLSCR1 is a cell-autonomous defence factor against SARS-CoV-2 infection
Abstract
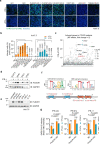
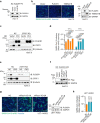
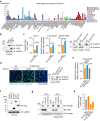
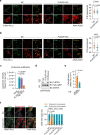
Understanding protective immunity to COVID-19 facilitates preparedness for future pandemics and combats new SARS-CoV-2 variants emerging in the human population. Neutralizing antibodies have been widely studied; however, on the basis of large-scale exome sequencing of protected versus severely ill patients with COVID-19, local cell-autonomous defence is also crucial1-4. Here we identify phospholipid scramblase 1 (PLSCR1) as a potent cell-autonomous restriction factor against live SARS-CoV-2 infection in parallel genome-wide CRISPR-Cas9 screens of human lung epithelia and hepatocytes before and after stimulation with interferon-γ (IFNγ). IFNγ-induced PLSCR1 not only restricted SARS-CoV-2 USA-WA1/2020, but was also effective against the Delta B.1.617.2 and Omicron BA.1 lineages. Its robust activity extended to other highly pathogenic coronaviruses, was functionally conserved in bats and mice, and interfered with the uptake of SARS-CoV-2 in both the endocytic and the TMPRSS2-dependent fusion routes. Whole-cell 4Pi single-molecule switching nanoscopy together with bipartite nano-reporter assays found that PLSCR1 directly targeted SARS-CoV-2-containing vesicles to prevent spike-mediated fusion and viral escape. A PLSCR1 C-terminal β-barrel domain-but not lipid scramblase activity-was essential for this fusogenic blockade. Our mechanistic studies, together with reports that COVID-associated PLSCR1 mutations are found in some susceptible people3,4, identify an anti-coronavirus protein that interferes at a late entry step before viral RNA is released into the host-cell cytosol.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
PLSCR1 suppresses SARS-CoV-2 infection by downregulating cell surface ACE2.J Virol. 2025 Mar 18;99(3):e0208524. doi: 10.1128/jvi.02085-24. Epub 2025 Feb 13. J Virol. 2025. PMID: 39945535 Free PMC article.
-
Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity.Nature. 2022 Mar;603(7902):706-714. doi: 10.1038/s41586-022-04474-x. Epub 2022 Feb 1. Nature. 2022. PMID: 35104837 Free PMC article.
-
Pan-beta-coronavirus subunit vaccine prevents SARS-CoV-2 Omicron, SARS-CoV, and MERS-CoV challenge.J Virol. 2024 Sep 17;98(9):e0037624. doi: 10.1128/jvi.00376-24. Epub 2024 Aug 27. J Virol. 2024. PMID: 39189731 Free PMC article.
-
Therapeutic nanobodies against SARS-CoV-2 and other pathogenic human coronaviruses.J Nanobiotechnology. 2024 May 31;22(1):304. doi: 10.1186/s12951-024-02573-7. J Nanobiotechnology. 2024. PMID: 38822339 Free PMC article. Review.
-
Phospholipid scramblase 1: a frontline defense against viral infections.Front Cell Infect Microbiol. 2025 Apr 3;15:1573373. doi: 10.3389/fcimb.2025.1573373. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40248364 Free PMC article. Review.
Cited by
-
Photocatalytic Microenvironment Proteomics of Thiol-Mediated Uptake.JACS Au. 2025 Jul 1;5(7):3288-3298. doi: 10.1021/jacsau.5c00432. eCollection 2025 Jul 28. JACS Au. 2025. PMID: 40747020 Free PMC article.
-
Signals of positive selection in genomes of palearctic Myotis-bats coexisting with a fungal pathogen.BMC Genomics. 2024 Sep 3;25(1):828. doi: 10.1186/s12864-024-10722-3. BMC Genomics. 2024. PMID: 39227786 Free PMC article.
-
Deciphering the molecular nexus between Omicron infection and acute kidney injury: a bioinformatics approach.Front Mol Biosci. 2024 Jul 4;11:1340611. doi: 10.3389/fmolb.2024.1340611. eCollection 2024. Front Mol Biosci. 2024. PMID: 39027131 Free PMC article.
-
Phospholipid Scramblase 1 (PLSCR1) Regulates Interferon-Lambda Receptor 1 (IFN-λR1) and IFN-λ Signaling in Influenza A Virus (IAV) Infection.bioRxiv [Preprint]. 2024 Nov 21:2024.11.20.624469. doi: 10.1101/2024.11.20.624469. bioRxiv. 2024. PMID: 39605457 Free PMC article. Preprint.
-
Pathological Glucose Levels Enhance Entry Factor Expression and Hepatic SARS-CoV-2 Infection.J Cell Mol Med. 2025 Jun;29(11):e70581. doi: 10.1111/jcmm.70581. J Cell Mol Med. 2025. PMID: 40442985 Free PMC article.
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

