Large language models encode clinical knowledge
- PMID: 37438534
- PMCID: PMC10396962
- DOI: 10.1038/s41586-023-06291-2
Large language models encode clinical knowledge
Erratum in
-
Publisher Correction: Large language models encode clinical knowledge.Nature. 2023 Aug;620(7973):E19. doi: 10.1038/s41586-023-06455-0. Nature. 2023. PMID: 37500979 Free PMC article. No abstract available.
Abstract
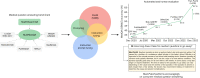
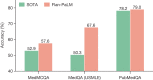
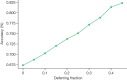
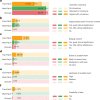
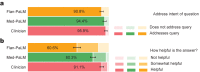
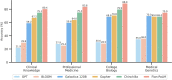
Large language models (LLMs) have demonstrated impressive capabilities, but the bar for clinical applications is high. Attempts to assess the clinical knowledge of models typically rely on automated evaluations based on limited benchmarks. Here, to address these limitations, we present MultiMedQA, a benchmark combining six existing medical question answering datasets spanning professional medicine, research and consumer queries and a new dataset of medical questions searched online, HealthSearchQA. We propose a human evaluation framework for model answers along multiple axes including factuality, comprehension, reasoning, possible harm and bias. In addition, we evaluate Pathways Language Model1 (PaLM, a 540-billion parameter LLM) and its instruction-tuned variant, Flan-PaLM2 on MultiMedQA. Using a combination of prompting strategies, Flan-PaLM achieves state-of-the-art accuracy on every MultiMedQA multiple-choice dataset (MedQA3, MedMCQA4, PubMedQA5 and Measuring Massive Multitask Language Understanding (MMLU) clinical topics6), including 67.6% accuracy on MedQA (US Medical Licensing Exam-style questions), surpassing the prior state of the art by more than 17%. However, human evaluation reveals key gaps. To resolve this, we introduce instruction prompt tuning, a parameter-efficient approach for aligning LLMs to new domains using a few exemplars. The resulting model, Med-PaLM, performs encouragingly, but remains inferior to clinicians. We show that comprehension, knowledge recall and reasoning improve with model scale and instruction prompt tuning, suggesting the potential utility of LLMs in medicine. Our human evaluations reveal limitations of today's models, reinforcing the importance of both evaluation frameworks and method development in creating safe, helpful LLMs for clinical applications.
© 2023. The Author(s).
Conflict of interest statement
This study was funded by Alphabet Inc. and/or a subsidiary thereof (Alphabet). K.S., S.A., T.T., V.N., A.K., S.S.M., C.S., J.W., H.W.C., N. Scales, A.T., H.C.-L., S.P., P.P., M.S., P.G., C.K., A.B., N. Schärli, A.C., P.M., B.A.A., D.W., G.S.C., Y.M., K.C., J.G., A.R., N.T., J.B. and Y.L. are employees of Alphabet and may own stock as part of the standard compensation package. D.D.-F. is affiliated with the US National Library of Medicine.
Figures


References
-
- Chowdhery, A. et al. PaLM: scaling language modeling with pathways. Preprint at 10.48550/arXiv.2204.02311 (2022).
-
- Chung, H. W. et al. Scaling instruction-finetuned language models. Preprint at 10.48550/arXiv.2210.11416 (2022).
-
- Jin, D. et al. What disease does this patient have? A large-scale open domain question answering dataset from medical exams. Appl. Sci.11, 6421 (2021).
-
- Pal, A., Umapathi, L. K. & Sankarasubbu, M. MedMCQA: a large-scale multi-subject multi-choice dataset for medical domain question answering. In Conference on Health, Inference, and Learning 248–260 (Proceedings of Machine Learning Research, 2022).
-
- Jin, Q., Dhingra, B., Liu, Z., Cohen, W. W. & Lu, X. PubMedQA: a dataset for biomedical research question answering. Preprint at 10.48550/arXiv.1909.06146 (2019).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

