N6-methyladenosine hypomethylation of circGPATCH2L regulates DNA damage and apoptosis through TRIM28 in intervertebral disc degeneration
- PMID: 37438603
- PMCID: PMC10406905
- DOI: 10.1038/s41418-023-01190-5
N6-methyladenosine hypomethylation of circGPATCH2L regulates DNA damage and apoptosis through TRIM28 in intervertebral disc degeneration
Abstract
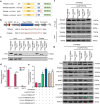
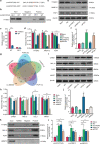
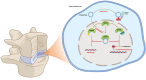
Circular RNAs (circRNAs) are a class of noncoding RNAs that have been found to be involved in intervertebral disc degeneration (IVDD) progression, and N6-methyladenosine (m6A) broadly exists in circRNAs. Here, we identified circGPATCH2L with a low m6A methylation level to be upregulated in degenerative nucleus pulposus tissues. Mechanistically, as a protein decoy for tripartite motif containing 28 (TRIM28) within aa 402-452 region, circGPATCH2L abrogates the phosphorylation of TRIM28 and inhibits P53 degradation, which contributes to DNA damage accumulation and cellular apoptosis and leads to IVDD progression. Moreover, m6A-methylated circGPATCH2L is recognised and endoribonucleolytically cleaved by a YTHDF2-RPL10-RNase P/MRP complex to maintain the physiological state of nucleus pulposus cells. Thus, our data show the physiological significance of m6A modification in regulating circRNA abundance and provide a potentially effective therapeutic target for the treatment of IVDD.
© 2023. The Author(s), under exclusive licence to ADMC Associazione Differenziamento e Morte Cellulare.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

