Identification of DraRS in Clostridioides difficile, a Two-Component Regulatory System That Responds to Lipid II-Interacting Antibiotics
- PMID: 37439672
- PMCID: PMC10601625
- DOI: 10.1128/jb.00164-23
Identification of DraRS in Clostridioides difficile, a Two-Component Regulatory System That Responds to Lipid II-Interacting Antibiotics
Abstract
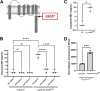
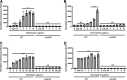
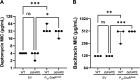
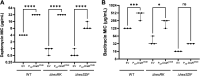
Clostridioides difficile is a Gram-positive opportunistic pathogen that results in 220,000 infections, 12,000 deaths, and upwards of $1 billion in medical costs in the United States each year. C. difficile is highly resistant to a variety of antibiotics, but we have a poor understanding of how C. difficile senses and responds to antibiotic stress and how such sensory systems affect clinical outcomes. We have identified a spontaneous C. difficile mutant that displays increased daptomycin resistance. We performed whole-genome sequencing and found a nonsense mutation, S605*, in draS, which encodes a putative sensor histidine kinase of a two-component system (TCS). The draSS605* mutant has an ~4- to 8-fold increase in the daptomycin MIC compared to the wild type (WT). We found that the expression of constitutively active DraRD54E in the WT increases daptomycin resistance 8- to 16-fold and increases bacitracin resistance ~4-fold. We found that a selection of lipid II-inhibiting compounds leads to the increased activity of the luciferase-based reporter PdraR-slucopt, including vancomycin, bacitracin, ramoplanin, and daptomycin. Using RNA sequencing (RNA-seq), we identified the DraRS regulon. Interestingly, we found that DraRS can induce the expression of the previously identified hex locus required for the synthesis of a novel glycolipid produced in C. difficile. Our data suggest that the induction of the hex locus by DraR explains some, but not all, of the DraR-induced daptomycin and bacitracin resistance. IMPORTANCE Clostridioides difficile is a major cause of hospital-acquired diarrhea and represents an urgent concern due to the prevalence of antibiotic resistance and the rate of recurrent infections. C. difficile encodes ~50 annotated two-component systems (TCSs); however, only a few have been studied. The function of these unstudied TCSs is not known. Here, we show that the TCS DraRS plays a role in responding to a subset of lipid II-inhibiting antibiotics and mediates resistance to daptomycin and bacitracin in part by inducing the expression of the recently identified hex locus, which encodes enzymes required for the production of a novel glycolipid in C. difficile.
Keywords: cell envelope; gene expression; signal transduction; stress response; two-component regulatory system; two-component regulatory systems.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Comment in
-
Expanding our grasp of two-component signaling in Clostridioides difficile.J Bacteriol. 2023 Oct 26;205(10):e0018823. doi: 10.1128/jb.00188-23. Epub 2023 Sep 20. J Bacteriol. 2023. PMID: 37728603 Free PMC article.
Similar articles
-
Expanding our grasp of two-component signaling in Clostridioides difficile.J Bacteriol. 2023 Oct 26;205(10):e0018823. doi: 10.1128/jb.00188-23. Epub 2023 Sep 20. J Bacteriol. 2023. PMID: 37728603 Free PMC article.
-
Identification of Clostridioides difficile mutants with increased daptomycin resistance.J Bacteriol. 2024 Mar 21;206(3):e0036823. doi: 10.1128/jb.00368-23. Epub 2024 Feb 20. J Bacteriol. 2024. PMID: 38376203 Free PMC article.
-
HexSDF Is Required for Synthesis of a Novel Glycolipid That Mediates Daptomycin and Bacitracin Resistance in C. difficile.mBio. 2023 Apr 25;14(2):e0339722. doi: 10.1128/mbio.03397-22. Epub 2023 Feb 14. mBio. 2023. PMID: 36786594 Free PMC article.
-
Clostridioides difficile - phage relationship the RNA way.Curr Opin Microbiol. 2022 Apr;66:1-10. doi: 10.1016/j.mib.2021.11.012. Epub 2021 Dec 15. Curr Opin Microbiol. 2022. PMID: 34922145 Review.
-
In vivo emergence of a still uncommon resistance to fidaxomicin in the urgent antimicrobial resistance threat Clostridioides difficile.J Antimicrob Chemother. 2023 Aug 2;78(8):1992-1999. doi: 10.1093/jac/dkad194. J Antimicrob Chemother. 2023. PMID: 37352110 Review.
Cited by
-
Expanding our grasp of two-component signaling in Clostridioides difficile.J Bacteriol. 2023 Oct 26;205(10):e0018823. doi: 10.1128/jb.00188-23. Epub 2023 Sep 20. J Bacteriol. 2023. PMID: 37728603 Free PMC article.
-
Identification of Clostridioides difficile mutants with increased daptomycin resistance.J Bacteriol. 2024 Mar 21;206(3):e0036823. doi: 10.1128/jb.00368-23. Epub 2024 Feb 20. J Bacteriol. 2024. PMID: 38376203 Free PMC article.
-
Histidine kinase-mediated cross-regulation of the vancomycin-resistance operon in Clostridioides difficile.Mol Microbiol. 2024 Jun;121(6):1182-1199. doi: 10.1111/mmi.15273. Epub 2024 May 1. Mol Microbiol. 2024. PMID: 38690761 Free PMC article.
References
-
- Centers for Disease Control and Prevention. 2019. Antibiotic resistance threats in the United States, 2019. Centers for Disease Control and Prevention, Atlanta, GA. https://stacks.cdc.gov/view/cdc/82532. Retrieved 4 January 2022.
-
- Hanberger H, Nilsson LE, Maller R, Isaksson B. 1991. Pharmacodynamics of daptomycin and vancomycin on Enterococcus faecalis and Staphylococcus aureus demonstrated by studies of initial killing and postantibiotic effect and influence of Ca2+ and albumin on these drugs. Antimicrob Agents Chemother 35:1710–1716. doi:10.1128/AAC.35.9.1710. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

