Identification and characterization of protein interactions with the major Niemann-Pick type C disease protein in yeast reveals pathways of therapeutic potential
- PMID: 37440478
- PMCID: PMC10471228
- DOI: 10.1093/genetics/iyad129
Identification and characterization of protein interactions with the major Niemann-Pick type C disease protein in yeast reveals pathways of therapeutic potential
Abstract
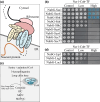
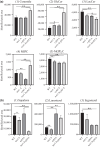
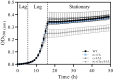
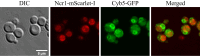
Niemann-Pick type C (NP-C) disease is a rare lysosomal storage disease caused by mutations in NPC1 (95% cases) or NPC2 (5% cases). These proteins function together in cholesterol egress from the lysosome, whereby upon mutation, cholesterol and other lipids accumulate causing major pathologies. However, it is not fully understood how cholesterol is transported from NPC1 residing at the lysosomal membrane to the endoplasmic reticulum (ER) and plasma membrane. The yeast ortholog of NPC1, Niemann-Pick type C-related protein-1 (Ncr1), functions similarly to NPC1; when transfected into a mammalian cell lacking NPC1, Ncr1 rescues the diagnostic hallmarks of cholesterol and sphingolipid accumulation. Here, we aimed to identify and characterize protein-protein interactions (PPIs) with the yeast Ncr1 protein. A genome-wide split-ubiquitin membrane yeast two-hybrid (MYTH) protein interaction screen identified 11 ER membrane-localized, full-length proteins interacting with Ncr1 at the lysosomal/vacuolar membrane. These highlight the importance of ER-vacuole membrane interface and include PPIs with the Cyb5/Cbr1 electron transfer system, the ceramide synthase complex, and the Sec61/Sbh1 protein translocation complex. These PPIs were not detected in a sterol auxotrophy condition and thus depend on normal sterol metabolism. To provide biological context for the Ncr1-Cyb5 PPI, a yeast strain lacking this PPI (via gene deletions) exhibited altered levels of sterols and sphingolipids including increased levels of glucosylceramide that mimic NP-C disease. Overall, the results herein provide new physical and genetic interaction models to further use the yeast model of NP-C disease to better understand human NP-C disease.
Keywords: lipid transport; lysosomal storage disease; neurodegenerative disease; rare disease; sphingolipid; sterol; yeast model.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest: The authors declare no conflict of interest.
Figures




Similar articles
-
An "exacerbate-reverse" strategy in yeast identifies histone deacetylase inhibition as a correction for cholesterol and sphingolipid transport defects in human Niemann-Pick type C disease.J Biol Chem. 2011 Jul 8;286(27):23842-51. doi: 10.1074/jbc.M111.227645. Epub 2011 Apr 13. J Biol Chem. 2011. PMID: 21489983 Free PMC article.
-
Mutagenesis of the putative sterol-sensing domain of yeast Niemann Pick C-related protein reveals a primordial role in subcellular sphingolipid distribution.J Cell Biol. 2004 Feb 16;164(4):547-56. doi: 10.1083/jcb.200310046. J Cell Biol. 2004. PMID: 14970192 Free PMC article.
-
Iron Limitation Restores Autophagy and Increases Lifespan in the Yeast Model of Niemann-Pick Type C1.Int J Mol Sci. 2023 Mar 25;24(7):6221. doi: 10.3390/ijms24076221. Int J Mol Sci. 2023. PMID: 37047194 Free PMC article.
-
Sterol uptake by the NPC system in eukaryotes: a Saccharomyces cerevisiae perspective.FEBS Lett. 2022 Jan;596(2):160-179. doi: 10.1002/1873-3468.14253. Epub 2021 Dec 20. FEBS Lett. 2022. PMID: 34897668 Review.
-
Finding pathogenic commonalities between Niemann-Pick type C and other lysosomal storage disorders: Opportunities for shared therapeutic interventions.Biochim Biophys Acta Mol Basis Dis. 2020 Oct 1;1866(10):165875. doi: 10.1016/j.bbadis.2020.165875. Epub 2020 Jun 6. Biochim Biophys Acta Mol Basis Dis. 2020. PMID: 32522631 Review.
Cited by
-
Structural and biochemical analysis of ligand binding in yeast Niemann-Pick type C1-related protein.Life Sci Alliance. 2024 Oct 25;8(1):e202402990. doi: 10.26508/lsa.202402990. Print 2025 Jan. Life Sci Alliance. 2024. PMID: 39455279 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

