Variability of the innate immune response is globally constrained by transcriptional bursting
- PMID: 37441161
- PMCID: PMC10333517
- DOI: 10.3389/fmolb.2023.1176107
Variability of the innate immune response is globally constrained by transcriptional bursting
Abstract
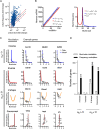
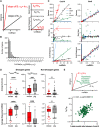
Transcription of almost all mammalian genes occurs in stochastic bursts, however the fundamental control mechanisms that allow appropriate single-cell responses remain unresolved. Here we utilise single cell genomics data and stochastic models of transcription to perform global analysis of the toll-like receptor (TLR)-induced gene expression variability. Based on analysis of more than 2000 TLR-response genes across multiple experimental conditions we demonstrate that the single-cell, gene-by-gene expression variability can be empirically described by a linear function of the population mean. We show that response heterogeneity of individual genes can be characterised by the slope of the mean-variance line, which captures how cells respond to stimulus and provides insight into evolutionary differences between species. We further demonstrate that linear relationships theoretically determine the underlying transcriptional bursting kinetics, revealing different regulatory modes of TLR response heterogeneity. Stochastic modelling of temporal scRNA-seq count distributions demonstrates that increased response variability is associated with larger and more frequent transcriptional bursts, which emerge via increased complexity of transcriptional regulatory networks between genes and different species. Overall, we provide a methodology relying on inference of empirical mean-variance relationships from single cell data and new insights into control of innate immune response variability.
Keywords: burst frequency; burst size; innate immunity; scRNA-seq inference; stochastic transcription; telegraph model; toll-like receptor; transcriptional bursting.
Copyright © 2023 Alachkar, Norton, Wolkensdorfer, Muldoon and Paszek.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Gene-Specific Linear Trends Constrain Transcriptional Variability of the Toll-like Receptor Signaling.Cell Syst. 2020 Sep 23;11(3):300-314.e8. doi: 10.1016/j.cels.2020.08.007. Epub 2020 Sep 11. Cell Syst. 2020. PMID: 32918862 Free PMC article.
-
BISC: accurate inference of transcriptional bursting kinetics from single-cell transcriptomic data.Brief Bioinform. 2022 Nov 19;23(6):bbac464. doi: 10.1093/bib/bbac464. Brief Bioinform. 2022. PMID: 36326081
-
Inferring transcriptional bursting kinetics from single-cell snapshot data using a generalized telegraph model.R Soc Open Sci. 2023 Apr 5;10(4):221057. doi: 10.1098/rsos.221057. eCollection 2023 Apr. R Soc Open Sci. 2023. PMID: 37035293 Free PMC article.
-
Transcriptional bursting dynamics in gene expression.Front Genet. 2024 Sep 13;15:1451461. doi: 10.3389/fgene.2024.1451461. eCollection 2024. Front Genet. 2024. PMID: 39346775 Free PMC article. Review.
-
What shapes eukaryotic transcriptional bursting?Mol Biosyst. 2017 Jun 27;13(7):1280-1290. doi: 10.1039/c7mb00154a. Mol Biosyst. 2017. PMID: 28573295 Review.
Cited by
-
Spatial microenvironments tune immune response dynamics in the Drosophila larval fat body.bioRxiv [Preprint]. 2024 Sep 16:2024.09.12.612587. doi: 10.1101/2024.09.12.612587. bioRxiv. 2024. PMID: 39345471 Free PMC article. Preprint.
References
-
- Akaike H. (1973). “Information theory and an extension of the maximum likelihood principle,” in Selected papers of hirotugu Akaike (Berlin, Germany: Springer; ).
-
- Al-Mohy A. H., Higham N. J. (2011). Computing the action of the matrix exponential, with an application to exponential integrators. Siam J. Sci. Comput. 33, 488–511. 10.1137/100788860 - DOI
LinkOut - more resources
Full Text Sources

