Bioactive silica nanoparticles target autophagy, NF-κB, and MAPK pathways to inhibit osteoclastogenesis
- PMID: 37441901
- PMCID: PMC10530178
- DOI: 10.1016/j.biomaterials.2023.122238
Bioactive silica nanoparticles target autophagy, NF-κB, and MAPK pathways to inhibit osteoclastogenesis
Abstract
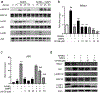
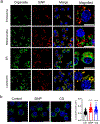
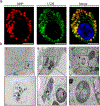
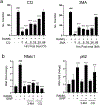
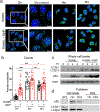
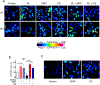
Spherical 50 nm silica-based nanoparticles (SiNPs) promote healthy bone homeostasis and maintenance by supporting bone forming osteoblast lineage cells while simultaneously inhibiting the differentiation of bone resorbing osteoclasts. Previous work demonstrated that an intraperitoneal injection of SiNPs in healthy mice - both young and old - increased bone density and quality, suggesting the possibility that SiNPs represent a dual action therapeutic. However, the underlying mechanisms governing the osteoclast response to SiNPs have yet to be fully explored and defined. Therefore, the goals of this study were to investigate the cellular and molecular mechanisms by which SiNPs inhibit osteoclastogenesis. SiNPs strongly inhibited RANKL-induced osteoclast differentiation within the first hours and concomitantly inhibited early transcriptional regulators such as Nfatc1. SiNPs simultaneously stimulated expression of autophagy related genes p62 and LC3β dependent on ERK1/2 signaling pathway. Intriguingly, SiNPs were found to stimulate autophagosome formation while inhibiting the autophagic flux necessary for RANKL-stimulated osteoclast differentiation, resulting in the inhibition of both the canonical and non-canonical NF-κB signaling pathways and stabilizing TRAF3. These results suggest a model in which SiNPs inhibit osteoclastogenesis by inhibiting the autophagic machinery and RANKL-dependent functionality. This mechanism of action defines a novel therapeutic strategy for inhibiting osteoclastogenesis.
Keywords: Autophagy; Bone resorption; LC3β; Nfatc1; Osteoporosis; p62.
Published by Elsevier Ltd.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





References
-
- Beck GR Jr., Ha SW, Camalier CE, Yamaguchi M, Li Y, Lee JK, Weitzmann MN, Bioactive silicabased nanoparticles stimulate bone-forming osteoblasts, suppress bone-resorbing osteoclasts, and enhance bone mineral density in vivo, Nanomedicine: nanotechnology, biology, and medicine 8(6) (2012) 793–803. - PMC - PubMed
-
- Udagawa N, Takahashi N, Akatsu T, Tanaka H, Sasaki T, Nishihara T, Koga T, Martin TJ, Suda T, Origin of osteoclasts: mature monocytes and macrophages are capable of differentiating into osteoclasts under a suitable microenvironment prepared by bone marrow-derived stromal cells, Proc Natl Acad Sci U S A 87(18) (1990) 7260–4. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

