An HBV susceptibility variant of KNG1 modulates the therapeutic effects of interferons α and λ1 in HBV infection by promoting MAVS lysosomal degradation
- PMID: 37442062
- PMCID: PMC10435766
- DOI: 10.1016/j.ebiom.2023.104694
An HBV susceptibility variant of KNG1 modulates the therapeutic effects of interferons α and λ1 in HBV infection by promoting MAVS lysosomal degradation
Abstract
Background: Hepatitis B virus (HBV) infection is one of the main causes of hepatocellular carcinoma (HCC). The relationship between HBV infection and the host genome as well as their underlying mechanisms remain largely unknown.
Methods: In this study, we performed a whole-genome exon sequencing analysis of 300 sib-pairs of Chinese HBV-infected families with the goal of identifying variants and genes involved in HBV infection. A site-direct mutant plasmid was used to investigate the function of SNP rs76438938 in KNG1. The functional and mechanical studies of KNG1 were conducted with in vitro liver cell lines and a hydrodynamic injection model in vivo. The impact of KNG1 on HBV infection therapy was determined in hepatocytes treated with IFN-α/λ1.
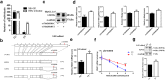
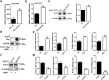
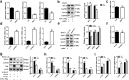
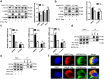
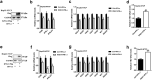
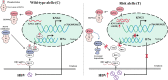
Findings: Our whole-exon association study of 300 families with hepatitis B infection found that SNP rs76438938 in KNG1 significantly increased the risk for HBV infection, and the rs76438938-T allele was found to promote HBV replication by increasing the stability of KNG1 mRNA. By competitively binding HSP90A with MAVS, KNG1 can inhibit the expression of types I and III IFNs by promoting MAVS lysosomal degradation. Such suppression of IFN expression and promotion of HBV replication by Kng1 were further demonstrated with an animal model in vivo. Lastly, we showed that the rs76438938-C allele can improve the therapeutic effect of IFN-α and -λ1 in HBV infection.
Interpretation: This study identified a SNP, rs76438938, in a newly discovered host gene, KNG1, for its involvement in HBV infection and treatment effect through modulating the cellular antiviral process.
Funding: This study was supported in part by the Independent Task of State Key Laboratory for Diagnosis and Treatment of Infectious Diseases of the First Affiliated Hospital of Zhejiang University, the China Precision Medicine Initiative (2016YFC0906300), and the Research Center for Air Pollution and Health of Zhejiang University.
Keywords: Functional SNP; HBV infection; Interferon; KNG1; Treatment.
Copyright © 2023 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests The authors report no biomedical financial interests or other potential conflicts of interest with the work reported in this paper.
Figures


Similar articles
-
Comparative Analysis of the Antiviral Effects Mediated by Type I and III Interferons in Hepatitis B Virus-Infected Hepatocytes.J Infect Dis. 2019 Jul 19;220(4):567-577. doi: 10.1093/infdis/jiz143. J Infect Dis. 2019. PMID: 30923817
-
Inducible interleukin 32 (IL-32) exerts extensive antiviral function via selective stimulation of interferon λ1 (IFN-λ1).J Biol Chem. 2013 Jul 19;288(29):20927-20941. doi: 10.1074/jbc.M112.440115. Epub 2013 May 31. J Biol Chem. 2013. PMID: 23729669 Free PMC article.
-
Interferon-inducible MX2 is a host restriction factor of hepatitis B virus replication.J Hepatol. 2020 May;72(5):865-876. doi: 10.1016/j.jhep.2019.12.009. Epub 2019 Dec 18. J Hepatol. 2020. PMID: 31863794
-
[Correlations between genetic polymorphism of IFN-λ family gene and HBV infection, virus replication and clearance].Sheng Wu Gong Cheng Xue Bao. 2022 Mar 25;38(3):893-902. doi: 10.13345/j.cjb.210310. Sheng Wu Gong Cheng Xue Bao. 2022. PMID: 35355462 Review. Chinese.
-
Interferon signaling during Hepatitis B Virus (HBV) infection and HBV-associated hepatocellular carcinoma.Cytokine. 2019 Dec;124:154518. doi: 10.1016/j.cyto.2018.08.012. Epub 2018 Aug 17. Cytokine. 2019. PMID: 30126685 Free PMC article. Review.
Cited by
-
Causal relationships between three plasma proteins and non-alcoholic fatty liver disease, mediated by Epstein-Barr virus EA-D antibody levels: a mendelian randomization study.Sci Rep. 2024 Oct 27;14(1):25644. doi: 10.1038/s41598-024-77105-2. Sci Rep. 2024. PMID: 39463412 Free PMC article.
-
Hepatitis B virus genotype distribution and mutation patterns: Insights and clinical implications for hepatitis B virus positive patients.World J Exp Med. 2025 Jun 20;15(2):102395. doi: 10.5493/wjem.v15.i2.102395. eCollection 2025 Jun 20. World J Exp Med. 2025. PMID: 40546674 Free PMC article. Review.
References
-
- Lai C.L., Ratziu V., Yuen M.F., Poynard T. Viral hepatitis B. Lancet. 2003;362(9401):2089–2094. - PubMed
-
- Perz J.F., Armstrong G.L., Farrington L.A., Hutin Y.J., Bell B.P. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45(4):529–538. - PubMed
-
- Polaris Observatory Collaborators Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet Gastroenterol Hepatol. 2018;3(6):383–403. - PubMed
-
- Ganem D., Prince A.M. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350(11):1118–1129. - PubMed
-
- Davila S., Froeling F.E., Tan A., et al. New genetic associations detected in a host response study to hepatitis B vaccine. Genes Immun. 2010;11(3):232–238. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

