Genetic inactivation of the Carnitine/Acetyl-Carnitine mitochondrial carrier of Yarrowia lipolytica leads to enhanced odd-chain fatty acid production
- PMID: 37443049
- PMCID: PMC10339547
- DOI: 10.1186/s12934-023-02137-8
Genetic inactivation of the Carnitine/Acetyl-Carnitine mitochondrial carrier of Yarrowia lipolytica leads to enhanced odd-chain fatty acid production
Abstract
Background: Mitochondrial carriers (MCs) can deeply affect the intracellular flux distribution of metabolic pathways. The manipulation of their expression level, to redirect the flux toward the production of a molecule of interest, is an attractive target for the metabolic engineering of eukaryotic microorganisms. The non-conventional yeast Yarrowia lipolytica is able to use a wide range of substrates. As oleaginous yeast, it directs most of the acetyl-CoA therefrom generated towards the synthesis of lipids, which occurs in the cytoplasm. Among them, the odd-chain fatty acids (OCFAs) are promising microbial-based compounds with several applications in the medical, cosmetic, chemical and agricultural industries.
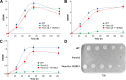
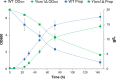
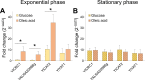
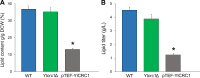
Results: In this study, we have identified the MC involved in the Carnitine/Acetyl-Carnitine shuttle in Y. lipolytica, YlCrc1. The Y. lipolytica Ylcrc1 knock-out strain failed to grow on ethanol, acetate and oleic acid, demonstrating the fundamental role of this MC in the transport of acetyl-CoA from peroxisomes and cytoplasm into mitochondria. A metabolic engineering strategy involving the deletion of YlCRC1, and the recombinant expression of propionyl-CoA transferase from Ralstonia eutropha (RePCT), improved propionate utilization and its conversion into OCFAs. These genetic modifications and a lipogenic medium supplemented with glucose and propionate as the sole carbon sources, led to enhanced accumulation of OCFAs in Y. lipolytica.
Conclusions: The Carnitine/Acetyl-Carnitine shuttle of Y. lipolytica involving YlCrc1, is the sole pathway for transporting peroxisomal or cytosolic acetyl-CoA to mitochondria. Manipulation of this carrier can be a promising target for metabolic engineering approaches involving cytosolic acetyl-CoA, as demonstrated by the effect of YlCRC1 deletion on OCFAs synthesis.
Keywords: Acetyl-CoA; Carnitine/Acetyl-Carnitine shuttle; Metabolic engineering; Mitochondrial carrier; Odd-chain fatty acids (OCFAs); Yarrowia lipolytica.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
- 2020SBNHLH_003/Ministero dell'Università e della Ricerca (MUR)
- 2020SBNHLH_003/Ministero dell'Università e della Ricerca (MUR)
- 2020SBNHLH_003/Ministero dell'Università e della Ricerca (MUR)
- 2020SBNHLH_003/Ministero dell'Università e della Ricerca (MUR)
- 2020SBNHLH_003/Ministero dell'Università e della Ricerca (MUR)
LinkOut - more resources
Full Text Sources
Research Materials

