LY6E is a pan-coronavirus restriction factor in the respiratory tract
- PMID: 37443277
- PMCID: PMC11234902
- DOI: 10.1038/s41564-023-01431-w
LY6E is a pan-coronavirus restriction factor in the respiratory tract
Abstract
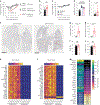
LY6E is an antiviral restriction factor that inhibits coronavirus spike-mediated fusion, but the cell types in vivo that require LY6E for protection from respiratory coronavirus infection are unknown. Here we used a panel of seven conditional Ly6e knockout mice to define which Ly6e-expressing cells confer control of airway infection by murine coronavirus and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Loss of Ly6e in Lyz2-expressing cells, radioresistant Vav1-expressing cells and non-haematopoietic cells increased susceptibility to murine coronavirus. Global conditional loss of Ly6e expression resulted in clinical disease and higher viral burden after SARS-CoV-2 infection, but little evidence of immunopathology. We show that Ly6e expression protected secretory club and ciliated cells from SARS-CoV-2 infection and prevented virus-induced loss of an epithelial cell transcriptomic signature in the lung. Our study demonstrates that lineage confined rather than broad expression of Ly6e sufficiently confers resistance to disease caused by murine and human coronaviruses.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures








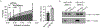
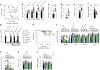


References
-
- Schoggins JW Interferon-stimulated genes: what do they all do? Annu. Rev. Virol 6, 567–584 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

