Transcription factor EGR2 controls homing and pathogenicity of TH17 cells in the central nervous system
- PMID: 37443284
- PMCID: PMC10500342
- DOI: 10.1038/s41590-023-01553-7
Transcription factor EGR2 controls homing and pathogenicity of TH17 cells in the central nervous system
Abstract
CD4+ T helper 17 (TH17) cells protect barrier tissues but also trigger autoimmunity. The mechanisms behind these opposing processes remain unclear. Here, we found that the transcription factor EGR2 controlled the transcriptional program of pathogenic TH17 cells in the central nervous system (CNS) but not that of protective TH17 cells at barrier sites. EGR2 was significantly elevated in myelin-reactive CD4+ T cells from patients with multiple sclerosis and mice with autoimmune neuroinflammation. The EGR2 transcriptional program was intricately woven within the TH17 cell transcriptional regulatory network and showed high interconnectivity with core TH17 cell-specific transcription factors. Mechanistically, EGR2 enhanced TH17 cell differentiation and myeloid cell recruitment to the CNS by upregulating pathogenesis-associated genes and myelomonocytic chemokines. T cell-specific deletion of Egr2 attenuated neuroinflammation without compromising the host's ability to control infections. Our study shows that EGR2 regulates tissue-specific and disease-specific functions in pathogenic TH17 cells in the CNS.
© 2023. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing Interest Statement
The authors declare no competing interests.
Figures


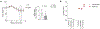

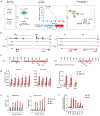

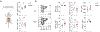



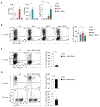






Comment in
-
EGR2 drives TH17 cell pathogenicity in autoimmune neuroinflammation.Nat Immunol. 2023 Aug;24(8):1230-1231. doi: 10.1038/s41590-023-01568-0. Nat Immunol. 2023. PMID: 37474655 No abstract available.
References
-
- Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL & Kuchroo VK Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238 (2006). - PubMed
-
- Mangan PR, Harrington LE, O’Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR & Weaver CT Transforming growth factor-beta induces development of the T(H)17 lineage. Nature 441, 231–234 (2006). - PubMed
-
- Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM & Stockinger B TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity 24, 179–189 (2006). - PubMed
-
- Ghoreschi K, Laurence A, Yang XP, Tato CM, McGeachy MJ, Konkel JE, Ramos HL, Wei L, Davidson TS, Bouladoux N, Grainger JR, Chen Q, Kanno Y, Watford WT, Sun HW, Eberl G, Shevach EM, Belkaid Y, Cua DJ, Chen W & O’Shea JJ Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature 467, 967–971 (2010). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

