The Troyer syndrome protein spartin mediates selective autophagy of lipid droplets
- PMID: 37443287
- PMCID: PMC10415183
- DOI: 10.1038/s41556-023-01178-w
The Troyer syndrome protein spartin mediates selective autophagy of lipid droplets
Abstract
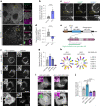
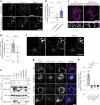
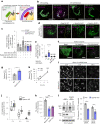
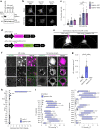
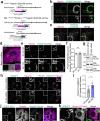
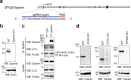
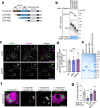
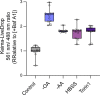
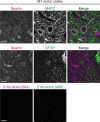
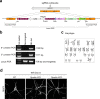
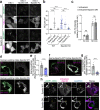
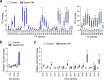
Lipid droplets (LDs) are crucial organelles for energy storage and lipid homeostasis. Autophagy of LDs is an important pathway for their catabolism, but the molecular mechanisms mediating LD degradation by selective autophagy (lipophagy) are unknown. Here we identify spartin as a receptor localizing to LDs and interacting with core autophagy machinery, and we show that spartin is required to deliver LDs to lysosomes for triglyceride mobilization. Mutations in SPART (encoding spartin) lead to Troyer syndrome, a form of complex hereditary spastic paraplegia1. Interfering with spartin function in cultured human neurons or murine brain neurons leads to LD and triglyceride accumulation. Our identification of spartin as a lipophagy receptor, thus, suggests that impaired LD turnover contributes to Troyer syndrome development.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Comment in
-
Spartin is a receptor for the autophagy of lipid droplets.Nat Cell Biol. 2023 Aug;25(8):1085-1086. doi: 10.1038/s41556-023-01201-0. Nat Cell Biol. 2023. PMID: 37474818 No abstract available.
-
SPART links autophagy machinery and lipid droplets in motor neurons.Autophagy. 2023 Nov;19(11):2835-2836. doi: 10.1080/15548627.2023.2247311. Epub 2023 Aug 22. Autophagy. 2023. PMID: 37573517 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

