Crosstalk between regulatory elements in disordered TRPV4 N-terminus modulates lipid-dependent channel activity
- PMID: 37443299
- PMCID: PMC10344929
- DOI: 10.1038/s41467-023-39808-4
Crosstalk between regulatory elements in disordered TRPV4 N-terminus modulates lipid-dependent channel activity
Abstract
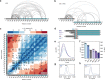
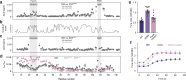
Intrinsically disordered regions (IDRs) are essential for membrane receptor regulation but often remain unresolved in structural studies. TRPV4, a member of the TRP vanilloid channel family involved in thermo- and osmosensation, has a large N-terminal IDR of approximately 150 amino acids. With an integrated structural biology approach, we analyze the structural ensemble of the TRPV4 IDR and the network of antagonistic regulatory elements it encodes. These modulate channel activity in a hierarchical lipid-dependent manner through transient long-range interactions. A highly conserved autoinhibitory patch acts as a master regulator by competing with PIP2 binding to attenuate channel activity. Molecular dynamics simulations show that loss of the interaction between the PIP2-binding site and the membrane reduces the force exerted by the IDR on the structured core of TRPV4. This work demonstrates that IDR structural dynamics are coupled to TRPV4 activity and highlights the importance of IDRs for TRP channel function and regulation.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








References
-
- Warnet XL, Bakke Krog H, Sevillano-Quispe OG, Poulsen H, Kjaergaard M. The C-terminal domains of the NMDA receptor: how intrinsically disordered tails affect signalling, plasticity and disease. Eur. J. Neurosci. 2021;54:6713–6739. - PubMed
-
- Goretzki B, Guhl C, Tebbe F, Harder J-M, Hellmich UA. Unstructural biology of TRP ion channels: the role of intrinsically disordered regions in channel function and regulation. J. Mol. Biol. 2021;433:166931. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

