Biophysical ordering transitions underlie genome 3D re-organization during cricket spermiogenesis
- PMID: 37443316
- PMCID: PMC10345107
- DOI: 10.1038/s41467-023-39908-1
Biophysical ordering transitions underlie genome 3D re-organization during cricket spermiogenesis
Abstract
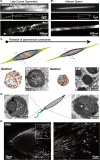
Spermiogenesis is a radical process of differentiation whereby sperm cells acquire a compact and specialized morphology to cope with the constraints of sexual reproduction while preserving their main cargo, an intact copy of the paternal genome. In animals, this often involves the replacement of most histones by sperm-specific nuclear basic proteins (SNBPs). Yet, how the SNBP-structured genome achieves compaction and accommodates shaping remain largely unknown. Here, we exploit confocal, electron and super-resolution microscopy, coupled with polymer modeling to identify the higher-order architecture of sperm chromatin in the needle-shaped nucleus of the emerging model cricket Gryllus bimaculatus. Accompanying spermatid differentiation, the SNBP-based genome is strikingly reorganized as ~25nm-thick fibers orderly coiled along the elongated nucleus axis. This chromatin spool is further found to achieve large-scale helical twisting in the final stages of spermiogenesis, favoring its ultracompaction. We reveal that these dramatic transitions may be recapitulated by a surprisingly simple biophysical principle based on a nucleated rigidification of chromatin linked to the histone-to-SNBP transition within a confined nuclear space. Our work highlights a unique, liquid crystal-like mode of higher-order genome organization in ultracompact cricket sperm, and establishes a multidisciplinary methodological framework to explore the diversity of non-canonical modes of DNA organization.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

