CSF MTBR-tau243 is a specific biomarker of tau tangle pathology in Alzheimer's disease
- PMID: 37443334
- PMCID: PMC10427417
- DOI: 10.1038/s41591-023-02443-z
CSF MTBR-tau243 is a specific biomarker of tau tangle pathology in Alzheimer's disease
Abstract
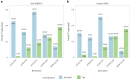
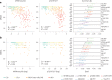
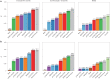
Aggregated insoluble tau is one of two defining features of Alzheimer's disease. Because clinical symptoms are strongly correlated with tau aggregates, drug development and clinical diagnosis need cost-effective and accessible specific fluid biomarkers of tau aggregates; however, recent studies suggest that the fluid biomarkers currently available cannot specifically track tau aggregates. We show that the microtubule-binding region (MTBR) of tau containing the residue 243 (MTBR-tau243) is a new cerebrospinal fluid (CSF) biomarker specific for insoluble tau aggregates and compared it to multiple other phosphorylated tau measures (p-tau181, p-tau205, p-tau217 and p-tau231) in two independent cohorts (BioFINDER-2, n = 448; and Knight Alzheimer Disease Research Center, n = 219). MTBR-tau243 was most strongly associated with tau-positron emission tomography (PET) and cognition, whereas showing the lowest association with amyloid-PET. In combination with p-tau205, MTBR-tau243 explained most of the total variance in tau-PET burden (0.58 ≤ R2 ≤ 0.75) and the performance in predicting cognitive measures (0.34 ≤ R2 ≤ 0.48) approached that of tau-PET (0.44 ≤ R2 ≤ 0.52). MTBR-tau243 levels longitudinally increased with insoluble tau aggregates, unlike CSF p-tau species. CSF MTBR-tau243 is a specific biomarker of tau aggregate pathology, which may be utilized in interventional trials and in the diagnosis of patients. Based on these findings, we propose to revise the A/T/(N) criteria to include MTBR-tau243 as representing insoluble tau aggregates ('T').
© 2023. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
K.H. is an Eisai-sponsored voluntary research associate professor at Washington University and has received salary from Eisai. Washington University. R.J.B. and D.M.H. have equity ownership interest in C2N Diagnostics. R.J.B. and D.M.H. receive income from C2N Diagnostics for serving on the scientific advisory board. K.H., N.R.B., C.S. and R.J.B. may receive income based on technology (Methods to Detect MTBR-tau Isoforms and use Thereof) (PCT/US2020/046224) licensed by Washington University to C2N Diagnostics. H.J. and D.M.H. may receive income based on technology (Anti-tau MTBR Antibodies and Methods to Detect Endogenously Cleaved Fragments of Tau and uses Thereof) (USSN 63/400,345) licensed by Washington University to C2N Diagnostics. R.J.B. is an unpaid scientific advisory board member of Roche and Biogen and receives research funding from Avid Radiopharmaceuticals, Janssen, Roche/Genentech, Eli Lilly, Eisai, Biogen, AbbVie, Bristol Myers Squibb and Novartis. O.H. has acquired research support (for the institution) from ADx, Avid Radiopharmaceuticals, Biogen, Eli Lilly, Eisai, Fujirebio, GE Healthcare, Pfizer and Roche. In the past 2 years, he has received consultancy/speaker fees from AC Immune, Amylyx, Alzpath, BioArctic, Biogen, Cerveau, Eisai, Fujirebio, Genentech, Novartis, Roche and Siemens. S.E.S. has analyzed data provided by C2N Diagnostics to Washington University, but she has not received any research funding or personal compensation from C2N Diagnostics. She has served on a scientific advisory board for Eisai. D.M.H. is on the scientific advisory board of Genentech, Denali and Cajal Neurosciences and consults for Alector. S.P. has served on scientific advisory boards and/or given lectures in symposia sponsored by BioArtic, Biogen, Cytox, Eli Lilly, Geras Solutions and Roche. The remaining authors declare no competing interests.
Figures


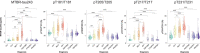



Comment in
-
A specific biomarker for insoluble tau.Nat Rev Drug Discov. 2023 Sep;22(9):697. doi: 10.1038/d41573-023-00126-5. Nat Rev Drug Discov. 2023. PMID: 37528207 No abstract available.
References
-
- Ossenkoppele R, van der Kant R, Hansson O. Tau biomarkers in Alzheimer’s disease: towards implementation in clinical practice and trials. Lancet Neurol. 2022;21:726–734. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

