Trichlorobacter ammonificans, a dedicated acetate-dependent ammonifier with a novel module for dissimilatory nitrate reduction to ammonia
- PMID: 37443340
- PMCID: PMC10504241
- DOI: 10.1038/s41396-023-01473-2
Trichlorobacter ammonificans, a dedicated acetate-dependent ammonifier with a novel module for dissimilatory nitrate reduction to ammonia
Erratum in
-
Correction: Trichlorobacter ammonificans, a dedicated acetate-dependent ammonifier with a novel module for dissimilatory nitrate reduction to ammonia.ISME J. 2024 Jan 8;18(1):wrad018. doi: 10.1093/ismejo/wrad018. ISME J. 2024. PMID: 38365262 Free PMC article. No abstract available.
Abstract
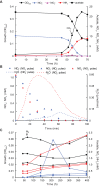
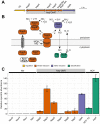
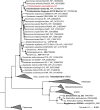
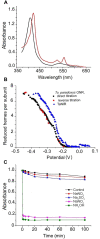
Dissimilatory nitrate reduction to ammonia (DNRA) is a common biochemical process in the nitrogen cycle in natural and man-made habitats, but its significance in wastewater treatment plants is not well understood. Several ammonifying Trichlorobacter strains (former Geobacter) were previously enriched from activated sludge in nitrate-limited chemostats with acetate as electron (e) donor, demonstrating their presence in these systems. Here, we isolated and characterized the new species Trichlorobacter ammonificans strain G1 using a combination of low redox potential and copper-depleted conditions. This allowed purification of this DNRA organism from competing denitrifiers. T. ammonificans is an extremely specialized ammonifier, actively growing only with acetate as e-donor and carbon source and nitrate as e-acceptor, but H2 can be used as an additional e-donor. The genome of G1 does not encode the classical ammonifying modules NrfAH/NrfABCD. Instead, we identified a locus encoding a periplasmic nitrate reductase immediately followed by an octaheme cytochrome c that is conserved in many Geobacteraceae species. We purified this octaheme cytochrome c protein (TaNiR), which is a highly active dissimilatory ammonifying nitrite reductase loosely associated with the cytoplasmic membrane. It presumably interacts with two ferredoxin subunits (NapGH) that donate electrons from the menaquinol pool to the periplasmic nitrate reductase (NapAB) and TaNiR. Thus, the Nap-TaNiR complex represents a novel type of highly functional DNRA module. Our results indicate that DNRA catalyzed by octaheme nitrite reductases is a metabolic feature of many Geobacteraceae, representing important community members in various anaerobic systems, such as rice paddy soil and wastewater treatment facilities.
© 2023. The Author(s), under exclusive licence to International Society for Microbial Ecology.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Dissimilatory Nitrate Reduction to Ammonium (DNRA) and Denitrification Pathways Are Leveraged by Cyclic AMP Receptor Protein (CRP) Paralogues Based on Electron Donor/Acceptor Limitation in Shewanella loihica PV-4.Appl Environ Microbiol. 2021 Jan 4;87(2):e01964-20. doi: 10.1128/AEM.01964-20. Print 2021 Jan 4. Appl Environ Microbiol. 2021. PMID: 33158888 Free PMC article.
-
Roles of NapF, NapG and NapH, subunits of the Escherichia coli periplasmic nitrate reductase, in ubiquinol oxidation.Mol Microbiol. 2002 Apr;44(1):245-55. doi: 10.1046/j.1365-2958.2002.02875.x. Mol Microbiol. 2002. PMID: 11967083
-
Unprecedented N2O production by nitrate-ammonifying Geobacteraceae with distinctive N2O isotopocule signatures.mBio. 2024 Dec 11;15(12):e0254024. doi: 10.1128/mbio.02540-24. Epub 2024 Oct 30. mBio. 2024. PMID: 39475233 Free PMC article.
-
Recent mechanistic developments for cytochrome c nitrite reductase, the key enzyme in the dissimilatory nitrate reduction to ammonium pathway.J Inorg Biochem. 2024 Jul;256:112542. doi: 10.1016/j.jinorgbio.2024.112542. Epub 2024 Mar 24. J Inorg Biochem. 2024. PMID: 38631103 Review.
-
Nitrate and periplasmic nitrate reductases.Chem Soc Rev. 2014 Jan 21;43(2):676-706. doi: 10.1039/c3cs60249d. Chem Soc Rev. 2014. PMID: 24141308 Free PMC article. Review.
Cited by
-
Adaptive responses of Trichlorobacter lovleyi to nitrite detoxification reveal overlooked contributions of Geobacterales to nitrate ammonification.ISME J. 2025 Jan 2;19(1):wraf054. doi: 10.1093/ismejo/wraf054. ISME J. 2025. PMID: 40101204 Free PMC article.
-
Growth physiology, genomics, and proteomics of Desulfurivibrio dismutans sp. nov., an obligately chemolithoautotrophic, sulfur disproportionating and ammonifying haloalkaliphile from soda lakes.Front Microbiol. 2025 May 23;16:1590477. doi: 10.3389/fmicb.2025.1590477. eCollection 2025. Front Microbiol. 2025. PMID: 40485835 Free PMC article.
-
Hydrogen-dependent dissimilatory nitrate reduction to ammonium enables growth of Campylobacterota isolates.ISME J. 2025 Jan 2;19(1):wraf092. doi: 10.1093/ismejo/wraf092. ISME J. 2025. PMID: 40367351 Free PMC article.
-
"Candidatus Siderophilus nitratireducens": a putative nap-dependent nitrate-reducing iron oxidizer within the new order Siderophiliales.ISME Commun. 2024 Jan 20;4(1):ycae008. doi: 10.1093/ismeco/ycae008. eCollection 2024 Jan. ISME Commun. 2024. PMID: 38577582 Free PMC article.
-
A novel mechanism for dissimilatory nitrate reduction to ammonium in Acididesulfobacillus acetoxydans.mSystems. 2024 Mar 19;9(3):e0096723. doi: 10.1128/msystems.00967-23. Epub 2024 Feb 7. mSystems. 2024. PMID: 38323850 Free PMC article.
References
-
- Behrendt A, de Beer D, Stief P. Vertical activity distribution of dissimilatory nitrate reduction in coastal marine sediments. Biogeosciences. 2013;10:7509–23. doi: 10.5194/bg-10-7509-2013. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

