Chromatin accessibility dynamics of neurogenic niche cells reveal defects in neural stem cell adhesion and migration during aging
- PMID: 37443352
- PMCID: PMC10353944
- DOI: 10.1038/s43587-023-00449-3
Chromatin accessibility dynamics of neurogenic niche cells reveal defects in neural stem cell adhesion and migration during aging
Abstract
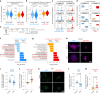
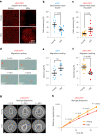
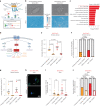
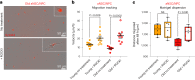
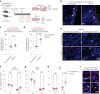
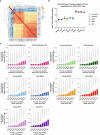
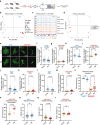
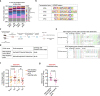
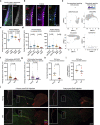
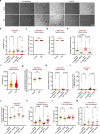
The regenerative potential of brain stem cell niches deteriorates during aging. Yet the mechanisms underlying this decline are largely unknown. Here we characterize genome-wide chromatin accessibility of neurogenic niche cells in vivo during aging. Interestingly, chromatin accessibility at adhesion and migration genes decreases with age in quiescent neural stem cells (NSCs) but increases with age in activated (proliferative) NSCs. Quiescent and activated NSCs exhibit opposing adhesion behaviors during aging: quiescent NSCs become less adhesive, whereas activated NSCs become more adhesive. Old activated NSCs also show decreased migration in vitro and diminished mobilization out of the niche for neurogenesis in vivo. Using tension sensors, we find that aging increases force-producing adhesions in activated NSCs. Inhibiting the cytoskeletal-regulating kinase ROCK reduces these adhesions, restores migration in old activated NSCs in vitro, and boosts neurogenesis in vivo. These results have implications for restoring the migratory potential of NSCs and for improving neurogenesis in the aged brain.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








References
-
- Gage FH, Temple S. Neural stem cells: generating and regenerating the brain. Neuron. 2013;80:588–601. - PubMed
-
- Silva-Vargas V, Crouch EE, Doetsch F. Adult neural stem cells and their niche: a dynamic duo during homeostasis, regeneration, and aging. Curr. Opin. Neurobiol. 2013;23:935–942. - PubMed
-
- Denoth-Lippuner A, Jessberger S. Formation and integration of new neurons in the adult hippocampus. Nat. Rev. Neurosci. 2021;22:223–236. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

