TOM1 family conservation within the plant kingdom for tobacco mosaic virus accumulation
- PMID: 37443447
- PMCID: PMC10576174
- DOI: 10.1111/mpp.13375
TOM1 family conservation within the plant kingdom for tobacco mosaic virus accumulation
Abstract
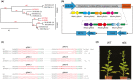
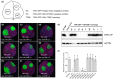
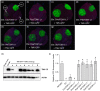
The susceptibility factor TOBAMOVIRUS MULTIPLICATION 1 (TOM1) is required for efficient multiplication of tobacco mosaic virus (TMV). Although some phylogenetic and functional analyses of the TOM1 family members have been conducted, a comprehensive analysis of the TOM1 homologues based on phylogeny from the most ancient to the youngest representatives within the plant kingdom, analysis of support for tobamovirus accumulation and interaction with other host and viral proteins has not been reported. In this study, using Nicotiana benthamiana and TMV as a model system, we functionally characterized the TOM1 homologues from N. benthamiana and other plant species from different plant lineages. We modified a multiplex genome editing tool and generated a sextuple mutant in which TMV multiplication was dramatically inhibited. We showed that TOM1 homologues from N. benthamiana exhibited variable capacities to support TMV multiplication. Evolutionary analysis revealed that the TOM1 family is restricted to the plant kingdom and probably originated in the Chlorophyta division, suggesting an ancient origin of the TOM1 family. We found that the TOM1 family acquired the ability to promote TMV multiplication after the divergence of moss and spikemoss. Moreover, the capacity of TOM1 orthologues from different plant species to promote TMV multiplication and the interactions between TOM1 and TOM2A and between TOM1 and TMV-encoded replication proteins are highly conserved, suggesting a conserved nature of the TOM2A-TOM1-TMV Hel module in promoting TMV multiplication. Our study not only revealed a conserved nature of a gene module to promote tobamovirus multiplication, but also provides a valuable strategy for TMV-resistant crop development.
Keywords: ToMV; evolution; genome editing; host factor; resistance; tobacco mosaic virus.
© 2023 The Authors. Molecular Plant Pathology published by British Society for Plant Pathology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Ali, M.E. , Ishii, Y. , Taniguchi, J.I. , Waliullah, S. , Kobayashi, K. , Yaeno, T. et al. (2018) Conferring virus resistance in tomato by independent RNA silencing of three tomato homologs of Arabidopsis TOM1 . Archives of Virology, 163, 1357–1362. - PubMed
-
- Asano, M. , Satoh, R. , Mochizuki, A. , Tsuda, S. , Yamanaka, T. , Nishiguchi, M. et al. (2005) Tobamovirus‐resistant tobacco generated by RNA interference directed against host genes. FEBS Letters, 579, 4479–4484. - PubMed
-
- Ding, X.S. , Liu, J. , Cheng, N.H. , Folimonov, A. , Hou, Y.M. , Bao, Y. et al. (2004) The tobacco mosaic virus 126‐kDa protein associated with virus replication and movement suppresses RNA silencing. Molecular Plant‐Microbe Interactions, 17, 583–592. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

