Tissue Inhibitor of Metalloproteinases-1 Overexpression Mediates Chemoresistance in Triple-Negative Breast Cancer Cells
- PMID: 37443843
- PMCID: PMC10340747
- DOI: 10.3390/cells12131809
Tissue Inhibitor of Metalloproteinases-1 Overexpression Mediates Chemoresistance in Triple-Negative Breast Cancer Cells
Abstract
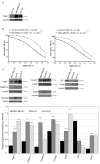
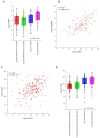
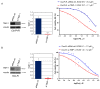
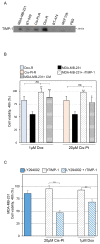
Triple-negative breast cancer (TNBC) is among the most aggressive breast cancer subtypes. Despite being initially responsive to chemotherapy, patients develop drug-resistant and metastatic tumors. Tissue inhibitor of metalloproteinases-1 (TIMP-1) is a secreted protein with a tumor suppressor function due to its anti-proteolytic activity. Nevertheless, evidence indicates that TIMP-1 binds to the CD63 receptor and activates noncanonical oncogenic signaling in several cancers, but its role in mediating TNBC chemoresistance is still largely unexplored. Here, we show that mesenchymal-like TNBC cells express TIMP-1, whose levels are further increased in cells generated to be resistant to cisplatin (Cis-Pt-R) and doxorubicin (Dox-R). Moreover, public dataset analyses indicate that high TIMP-1 levels are associated with a worse prognosis in TNBC subjected to chemotherapy. Knock-down of TIMP-1 in both Cis-Pt-R and Dox-R cells reverses their resistance by inhibiting AKT activation. Consistently, TNBC cells exposed to recombinant TIMP-1 or TIMP-1-enriched media from chemoresistant cells, acquire resistance to both cisplatin and doxorubicin. Importantly, released TIMP-1 reassociates with plasma membrane by binding to CD63 and, in the absence of CD63 expression, TIMP-1-mediated chemoresistance is blocked. Thus, our results identify TIMP-1 as a new biomarker of TNBC chemoresistance and lay the groundwork for evaluating whether blockade of TIMP-1 signal is a viable treatment strategy.
Keywords: CD63 cell receptor; chemoresistance; tissue inhibitor of metalloproteinases-1; triple-negative breast cancer; tumor microenvironment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

