RNA-Seq Analysis of Peripheral Whole Blood from Dairy Bulls with High and Low Antibody-Mediated Immune Responses-A Preliminary Study
- PMID: 37444006
- PMCID: PMC10339907
- DOI: 10.3390/ani13132208
RNA-Seq Analysis of Peripheral Whole Blood from Dairy Bulls with High and Low Antibody-Mediated Immune Responses-A Preliminary Study
Abstract
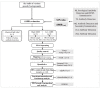
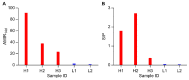
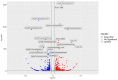
Enhancing the immune response through breeding is regarded as an effective strategy for improving animal health, as dairy cattle identified as high immune responders are reported to have a decreased prevalence of economically significant diseases. The identification of differentially expressed genes (DEGs) associated with immune responses might be an effective tool for breeding healthy dairy cattle. In this study, antibody-mediated immune responses (AMIRs) were induced by the immunization of hen egg white lysozyme (HEWL) in six Chinese Holstein dairy bulls divided into high- and low-AMIR groups based on their HEWL antibody level. Then, RNA-seq was applied to explore the transcriptome of peripheral whole blood between the two comparison groups. As a result, several major upregulated and downregulated genes were identified and attributed to the regulation of locomotion, tissue development, immune response, and detoxification. In addition, the result of the KEGG pathway analysis revealed that most DEGs were enriched in pathways related to disease, inflammation, and immune response, including antigen processing and presentation, Staphylococcus aureus infection, intestinal immune network for IgA production, cytokine-cytokine receptor interaction, and complement and coagulation cascades. Moreover, six genes (BOLA-DQA5, C5, CXCL2, HBA, LTF, and COL1A1) were validated using RT-qPCR, which may provide information for genomic selection in breeding programs. These results broaden the knowledge of the immune response mechanism in dairy bulls, which has strong implications for breeding cattle with an enhanced AMIR.
Keywords: RNA-seq; antibody-mediated immune response; dairy bulls; transcriptome analysis.
Conflict of interest statement
We certify that there are no conflict of interest with any financial organizations regarding the material discussed in the manuscript.
Figures
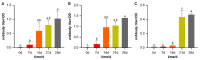





Similar articles
-
A genome-wide association study of immune response traits in Canadian Holstein cattle.BMC Genomics. 2014 Jul 4;15(1):559. doi: 10.1186/1471-2164-15-559. BMC Genomics. 2014. PMID: 24996426 Free PMC article.
-
Transcriptomic characterization of adult zebrafish infected with Streptococcus agalactiae.Fish Shellfish Immunol. 2019 Nov;94:355-372. doi: 10.1016/j.fsi.2019.09.040. Epub 2019 Sep 15. Fish Shellfish Immunol. 2019. PMID: 31533079
-
RNA-Seq transcriptomics and pathway analyses reveal potential regulatory genes and molecular mechanisms in high- and low-residual feed intake in Nordic dairy cattle.BMC Genomics. 2017 Mar 24;18(1):258. doi: 10.1186/s12864-017-3622-9. BMC Genomics. 2017. PMID: 28340555 Free PMC article.
-
Differences in udder health and immune response traits of Holstein-Friesians, Norwegian Reds, and their crosses in second lactation.J Dairy Sci. 2009 Feb;92(2):749-57. doi: 10.3168/jds.2008-1356. J Dairy Sci. 2009. PMID: 19164687
-
Review: Genomics of bull fertility.Animal. 2018 Jun;12(s1):s172-s183. doi: 10.1017/S1751731118000599. Epub 2018 Apr 5. Animal. 2018. PMID: 29618393 Free PMC article. Review.
References
-
- Oltenacu P., Broom D. The Impact of Genetic Selection for Increased Milk Yield on the Welfare of Dairy Cows. Anim. Welf. 2010;19:39–49. doi: 10.1017/S0962728600002220. - DOI
-
- Gaddis K.P. Development of National Genomic Evaluations for Health Traits in U.S. Holsteins; Proceedings of the World Congress of Genetics Applied in Livestock Production; Auckland, New Zealand. 11–16 February 2018; p. 594.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

